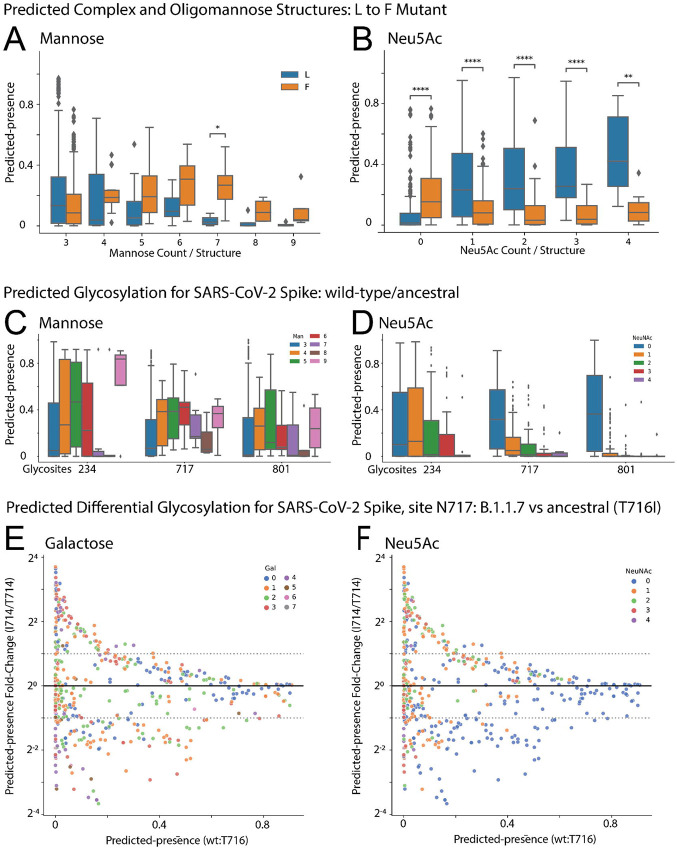
Figure 5. InSaNNE predicts complex glycans around the enhanced aromatic sequon and the SARS-CoV-2 spike protein.
(A-B) Boxplot distributions of predicted-presence for the L and F variants at N-2 stratified by number of (A) mannoses per glycan and (B) sialic acids per glycan. (C-D) Boxplots describing predicted glycosylation by (C) mannose per glycan and (D) sialic acid per glycan for three oligomannose sites in the SARS-CoV-2 spike glycoprotein. See Supplementary Figure 4 for all SARS-CoV-2 spike glycosylation sites. (E-F) Fold changes of predicted glycans at site N717, labeled by number of (E) galactose and (F) sialic acid units, between the wild-type and B.1.1.7 spike protein. Predicted-presence fold-change (y-axis) is stratified by the basal predicted-presence for each glycan in the wild-type (x-axis). Predicted-presence fold-change from wild-type by galactose, mannose, GlcNAc, and sialic acid is provided for N717 and N616 in B.1.1.7 (Supplementary Figure 5) and D615G (Supplementary Figure 6) variants respectively. ns: p>0.05, *: p<0.05, **: p<0.01, **: p<0.001, ***: p<1e-3, ****:p<1e-4