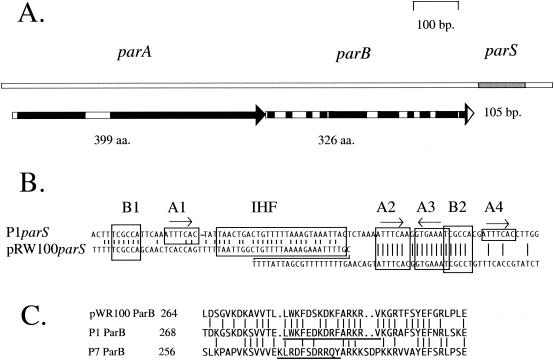
FIG. 1.
The pWR100 par region. A. Map of the pWR100par region showing the parA and parB open reading frames and the parS site. Black boxes on the open reading frame arrows show regions of very high similarity to the amino acid sequences of the equivalent P1 Par proteins. Blocks of >10 amino acids with >90% similarity to the P1 proteins are included. B. Alignment of the parS sequences of pWR100 and P1. The pWR100parS site is displayed in a staggered configuration in order to accommodate an additional 21-bp sequence that is absent from P1parS. Vertical lines join identities. The boxed sequences B1 and B2 are the discriminator hexamers whose variants determine the species specificity differences for parS between various P1par family members. The boxes A1 through A4 are repeats of the heptamer BoxA sequence with the consensus ATTTCAC. They are the principle binding motifs for the ParB protein. Boxes A1 and A4 are conserved in previously characterized P1par family members but appear to be absent in pWR100parS. C. Alignment of amino acid sequences from near the C termini of the ParB proteins of pWR100, P1, and P7. Vertical lines join identities. The underlined P1 and P7 sequences are the regions within which resides the information thought to be involved in the specific recognition of the respective P1 and P7parS BoxB motifs (20).