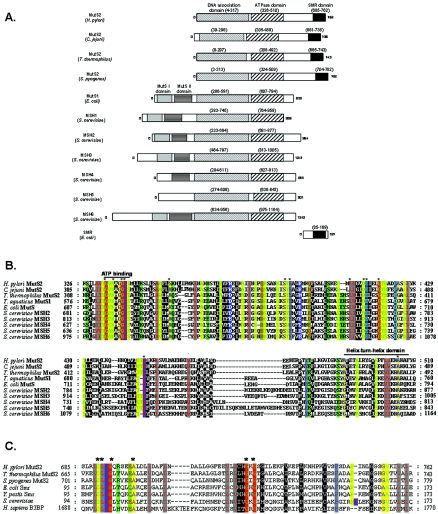
FIG. 1.
Genetic analyses of mutS1 and mutS2 homologs. A. Schematic of MutS homologs. All sequences were obtained from GenBank, and conserved protein domains were identified using PFam (26) and the SMART program (http://smart.embl-heidelberg.de) (42). B. Amino acid alignment of the ATPase domain of MutS homologs. The ATPase domain of MutS1 from E. coli and T. aquaticus, MSH2, -3, -4, and -6 from S. cerevisiae, and MutS2 from Listeria monocytogenes and H. pylori were aligned with ClustalX and visualized using the program Genedoc (www.psc.edu/biomed/genedoc) in physiochemical properties shading mode, which divides residues into 12 groups representing three hierarchies. The first hierarchy is based on size, the second is based electrical charge for the polar amino acids, and the third, for nonpolar amino acids, is based on aromaticity. The highly conserved ATP binding domain (73) and putative helix-turn-helix domain (12, 54) are indicated with a black line. C. Amino acid alignment of SMR domains. The SMR domain of H. pylori MutS2 was aligned with SMR domains from other MutS2 and Smr proteins. Also included were SMR domains from hypothetical proteins in Caenorhabditis elegans and Drosophila melanogaster and the human Bcl-3 binding protein (B3BP), which has recently been shown to possess nicking endonuclease activity (72). Alignments were constructed and visualized as described for panel B. Asterisks indicate residues with 100% conservation.