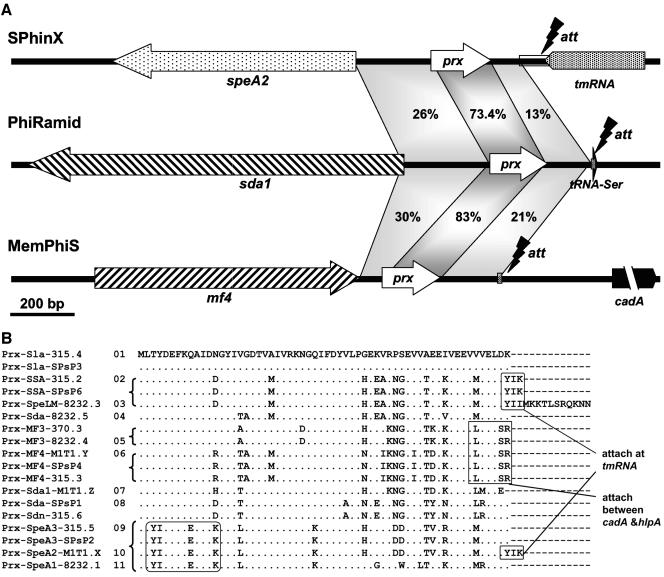
FIG. 4.
Paratox: a highly conserved ORF in M1T1 prophages. (A) A comparison between the lysogenic conversion modules and attachment sites of the three M1T1 prophages shows a highly conserved ORF that best matches a hypothetical phage protein located between each toxin and the phage attachment site. We named this hypothetical protein paratox (prx). Shaded areas indicate nucleotide similarity, and the percentage nucleotide identity is given. (B) Alignment of paratox protein alleles shows highly conserved amino acid sequence (represented by dots). Representative motifs linked to particular toxins or to phage attachment sites are boxed. All sequences are extracted from GenBank; in cases where the prx sequences were not annotated as ORFs, we picked them based on their similarity to the annotated ones. Each Prx will be referred to as (Prx_tox_Phi#), where tox is the name of the adjacent toxin and Phi# is the phage name and number (e.g., Prx_SpeA2_M1T1.X is the product of the paratox gene adjacent to SpeA2 in Phi M1T1.X, alias SPhinX). Serial numbers (1 to 11) were given to the distinct paratox alleles shown.