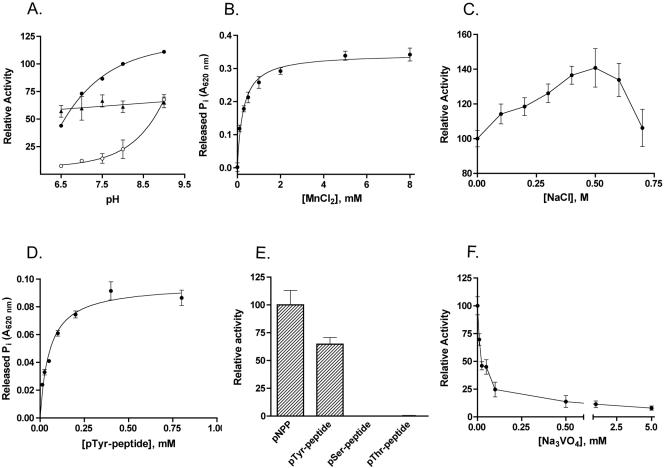
FIG. 1.
Biochemical characterization of recombinant YwqE. (A) Effect of pH on YwqE enzymatic activity in the presence of 1 mM MnCl2 (closed circles), 1 mM CuCl2 (triangles), or 1 mM ZnCl2 (open circles). The buffer used was 50 mM Tris-HCl. The graph shows datum points and regression. (B) Effect of MnCl2 on the activity of YwqE with pNPP as the substrate. The buffer used was 50 mM HEPES-KOH, pH 8.0, and contained increasing concentrations of MnCl2. The graph shows datum points and a nonlinear fit of the experimental data to the Michaelis-Menten equation. (C) Effect of NaCl on the activity of YwqE with pNPP as the substrate. (D) Analysis of YwqE enzymatic activity using the pTyr peptide KRLIEDNE(pY)TARGQ as a substrate. The graph shows datum points and a nonlinear fit of the experimental data to the Michaelis-Menten equation. (E) Comparison of the activities of YwqE using pNPP, the pTyr peptide KRLIEDNE(pY)TARGQ, the pSer peptide RRA(pS)VA, and the pThr peptide KR(pT)IRR as substrates. The assay mixture contained 0.5 mM pNPP or phosphopeptides as substrates. (F) Effect of Na3VO4 on the activity of YwqE with pNPP as the substrate. The buffer used for panels C and F was 50 mM HEPES-KOH, pH 8.0, and 1 mM MnCl2; the buffer used for panels D and E was 100 mM HEPES-KOH, pH 8.0, and 1 mM MnCl2. For panels A through C and F, the assay mixture contained 3 mM pNPP as a substrate. When error bars are not present in the graphs, they are within the resolution of the datum points.