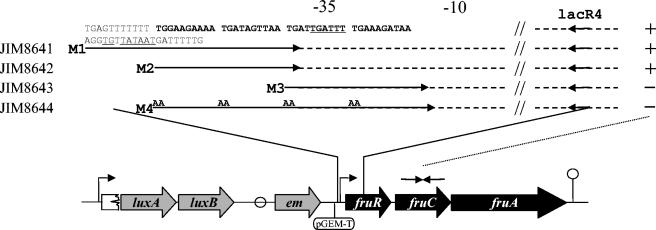
FIG. 4.
Schematic representation of the modified fru operon promoter sequences used to characterize the DNA-binding motif recognized by FruR. The promoter region inserted upstream of the fru operon is shown in the upper panel. The putative binding site of FruR is in bold. Strains were obtained by integration in the IL1403 chromosome of the plasmid pJIM2374 carrying pGEM-T, and the promoter regions amplified by the primers are indicated by arrows. Conserved sequences are shown by dotted lines, and TG→AA mutations in the M4 primer are indicated. The −10 and −35 regions are underlined. + or - at the right of the fragments indicate that the ratios of expression, as measured by QRT-PCR with lacC1 and lacC2 primers (complementary to the fruC gene) in cells grown in CDM with trehalose and fructose versus those grown in CDM-trehalose, are similar to that of the wild-type strain or close to 1, respectively.