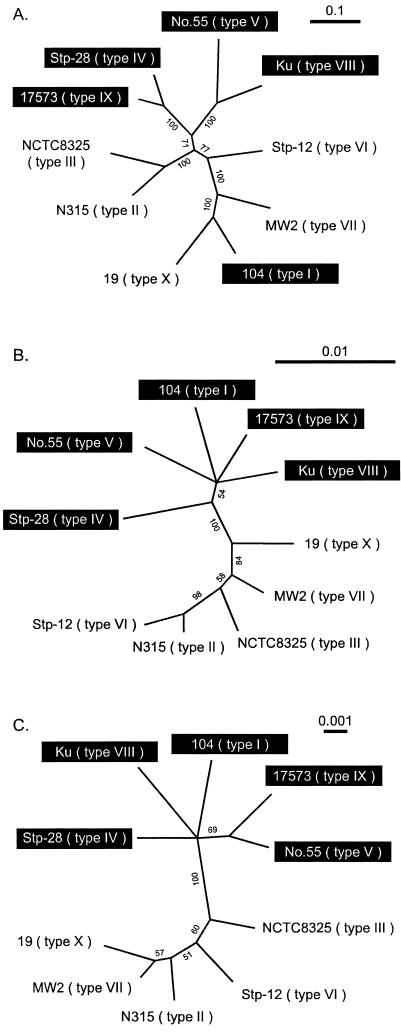
FIG. 3.
Maximum likelihood trees constructed using GCG paupsearch and visualized with Treeview software. Nucleotide sequences used for comparison were as follows: A, nucleotide sequencesof coa other than the repeat region and C-terminal sequence; B, regions surrounding coa which were concatenated sequences of SA0220 noncoding region 1, SA0221 noncoding region 2 on the left region flanked by coa and noncoding region 3, and SA0223 on the right region flanked by coa; C, housekeeping genes, which were concatenated parts of seven housekeeping genes used for MLST. Branches with significant bootstrap support (more than 50%) are indicated. Group 1 strains are highlighted by black boxes; the remaining are group 2 strains.