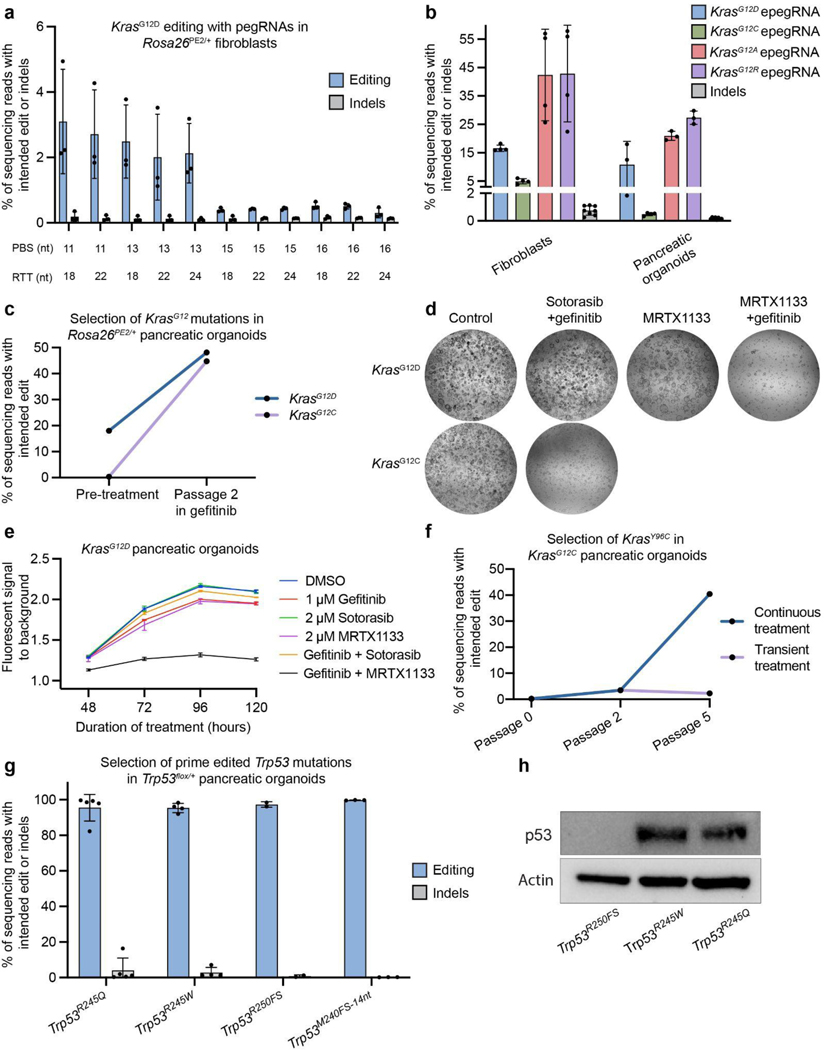
Figure 3.
Ex vivo prime editing and functional testing of Kras and Trp53 mutations.
a. Editing efficiency and indel byproduct frequency of the KrasG12D transition mutation (G:C to A:T) templated by a cohort of pegRNAs based on a single Cas9 spacer (n = 3 for each pegRNA). pegRNAs are delineated by differences in the lengths of the primer binding site (PBS) and reverse transcriptase template (RTT). Data and error bars indicate the mean and standard deviation of three independent transductions.
b. Editing activity of four engineered pegRNAs (epegRNAs) templating either the KrasG12D transition or the KrasG12C, KrasG12A, or KrasG12R transversions in tail tip-derived fibroblasts or pancreatic organoids (KrasG12D and KrasG12C). Data and error bars indicate the mean and standard deviation of three independent transductions. epegRNAs were generated by appending the trimmed evopreQ1 motif after the primer binding site of the leftmost pegRNA depicted in Fig. 1a. Indel byproduct calculations were pooled from all conditions within each tissue.
c. Allele frequencies of KrasG12D or KrasG12C mutations in pancreatic organoids before and after two passages of treatment with gefitinib (1 μM) (n = 1). Gefitinib treatment selects for cells containing prime edited KrasG12D or KrasG12C mutations.
d. Bright-field images of prime edited KrasG12C or KrasG12D organoids treated for four days with either control DMSO, sotorasib (2 μM) and gefitinib (1 μM), MRTX1133 (5 μM), or MRTX1133 and gefitinib.
e. Viability of KrasG12D pancreatic organoids under various treatment conditions. Viability was quantified using the alamarBlue HS Cell Viability Reagent, which is metabolized into a fluorescent derivative in living cells. Bars represent the range across two independent replicates.
f. Allele frequency of KrasY96C in KrasG12C organoids during and after treatment with sotorasib (2 μM) and gefitinib (n = 1). After two passages, organoids were split into two groups, which included continued treatment (continuous treatment) in one group and removal of treatment in a second group (transient treatment).
g. Allele frequencies of three Trp53 mutations in Trp53flox/+ pancreatic organoids treated with nutlin-3a for three to five passages after transduction with UPEC vectors. Indel byproduct frequencies are also included. Note that the highest indel frequency depicted for Trp53R245W derives primarily from a scaffold insertion in a single replicate. Trp53R245Q and Trp53R245W are homologous to mutations commonly observed in pancreatic cancer patients (TP53R248Q and TP53R248W), as described in Supplementary Figure 8. Trp53R250FS denotes a dinucleotide deletion that induces a frameshift mutation. Trp53M240FS−14nt denotes a fourteen-nucleotide deletion. Data and error bars indicate the mean and standard deviation of two to five independent transductions.
h. Immunoblot indicating detectable levels of p53 protein in prime edited Trp53flox/R245Q and Trp53flox/R245W organoids and an absence of detectable protein in Trp53flox/R250FS organoids.