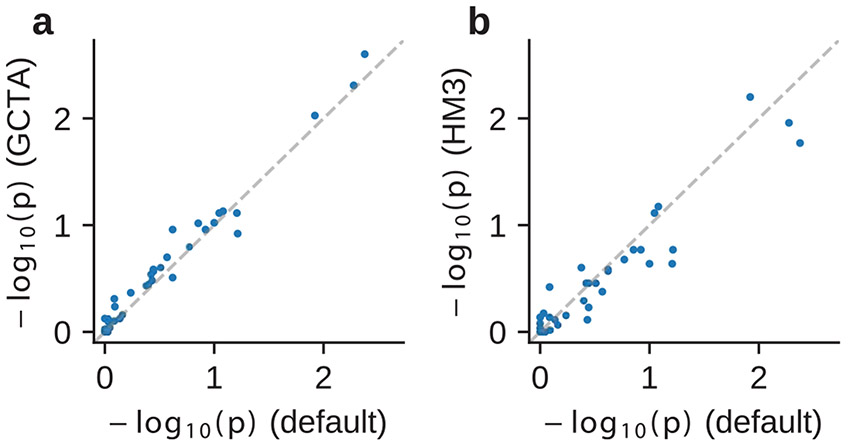
Extended Data Fig. 3 ∣. estimation is robust to genetic architecture and SNP set.
We performed estimation under the assumption of alternative genetic architecture and SNP set on real trait analysis across PAGE and UKBB. We compared -values (for one-sided test of ) of our default setting (using frequency-dependent genetic architecture and imputed SNPs; Table 1) to those obtained using GCTA genetic architecture and imputed SNPs (a), and to those obtained using frequency-dependent genetic architecture and HM3 SNPs (b). Numerical results are reported in Supplementary Table 8.