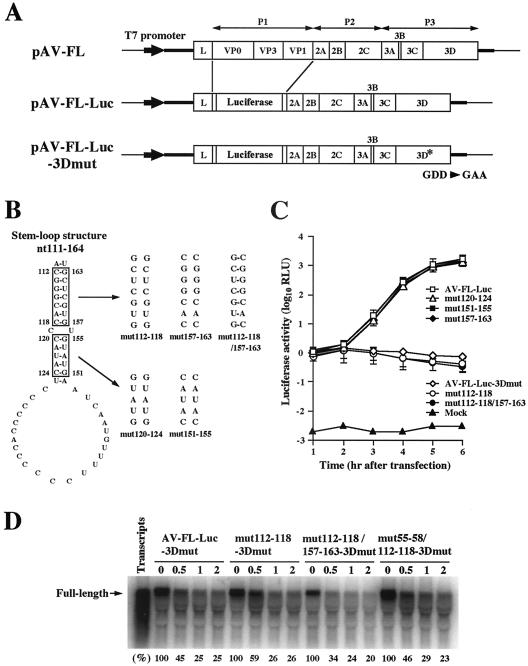
FIG. 2.
(A) Organization of the Aichi virus infectious cDNA clone, pAV-FL, and the Aichi virus replicon harboring a luciferase gene, pAV-FL-Luc, and pAV-FL-Luc-3Dmut. The thick lines and open boxes show the untranslated regions and coding regions, respectively. The thin lines indicate the vector sequences. The Aichi virus sequence was cloned just downstream of the T7 promoter sequence. (B) Site-directed mutations introduced into the stem-loop structure (nt 111 to 164) predicted downstream of SL-C. (C) Luciferase activity in cells transfected with AV-FL-Luc and mutants as to the predicted stem-loop structure. Vero cells were electroporated with the RNAs, and then at the indicated times after electroporation, cell lysates were prepared and analyzed for luciferase activity. The experiment was repeated at least three times. Standard deviation bars are shown. (D) RNA stability in Vero cell extracts. 32P-labeled RNA transcripts were incubated at 32°C. At the indicated times after incubation, RNA was extracted, treated with glyoxal, and analyzed by 1% agarose gel electrophoresis. The gel was dried, and radioactive signals were detected with a phosphorimager. Signal intensities of viral RNAs were quantitated, and the percentages of intensities relative to that obtained at 0 h after incubation are shown. Labeled AV-FL-Luc-3Dmut RNA was loaded in the lane represented as input RNA to show its electrophoretic mobility.