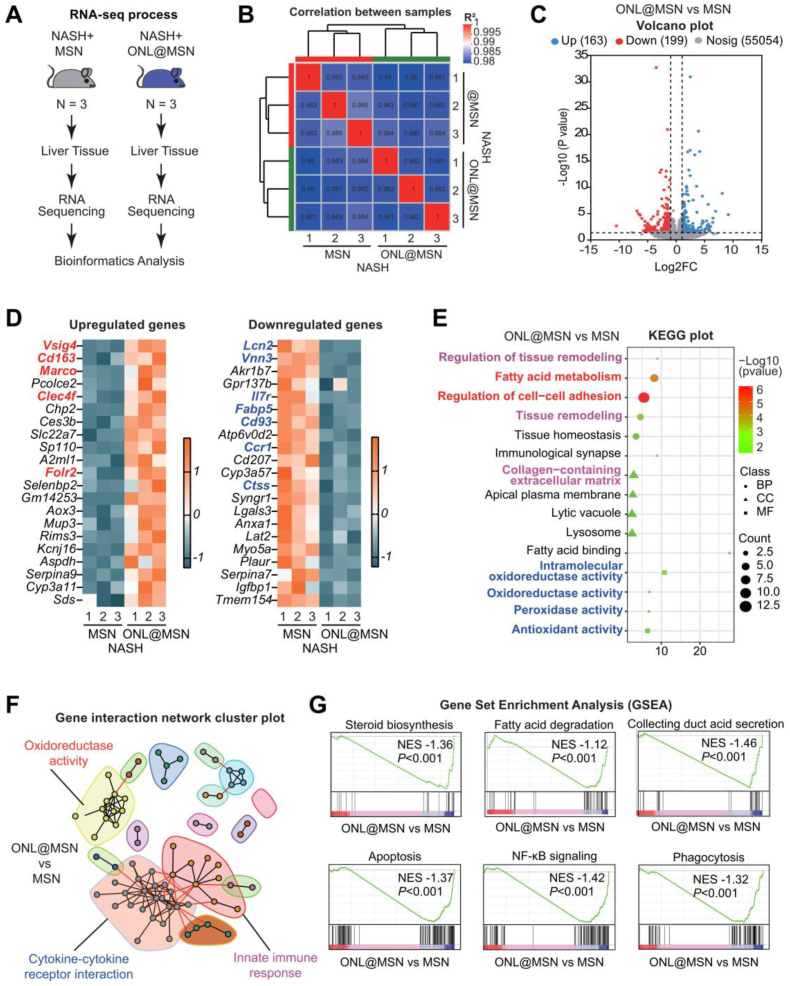
Figure 8.
Bulk-tissue RNA sequencing reveals the effects of ONL@MSN on the liver tissue transcriptome in NASH mice. (A) The experimental design involved performing bulk-tissue RNA sequencing on liver tissues from mice treated with ONL@MSN or MSN (control). (B) The correlation between samples from the two groups was assessed. (C) A volcano plot was generated to display the upregulated DEGs (n = 163) and downregulated DEGs (n = 199) in the liver tissue of mice treated with ONL@MSN compared to mice treated with MSN. (D) A heatmap plot was generated to show the top 20 upregulated and downregulated DEGs, respectively. (E) A KEGG plot was created to illustrate the main processes altered by ONL@MSN compared to MSN in terms of molecular function (MF), biological process (BP), and cell component (CC). (F) A gene interaction network cluster plot was generated to display the major important modules interacting with other genes. (G) A Gene Set Enrichment Analysis (GSEA) plot was generated to demonstrate several enriched signaling pathways resulting from the treatment of ONL@MSN compared to MSN.