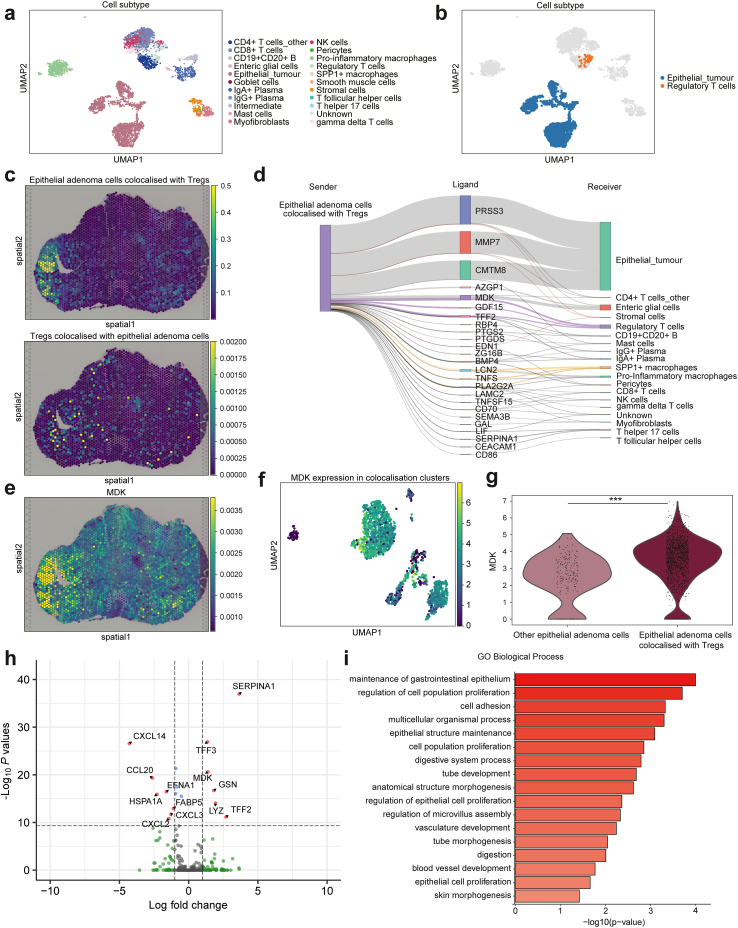
Fig. 3.
Colocalisation analysis and cell–cell interaction of adenoma epithelial cells and regulatory T cells. (a) Uniform manifold approximation and projection (UMAP) of cell subtypes in adenoma cluster. (b) Colocalisation clusters between adenoma epithelial cells and regulatory T cells (Tregs) in UMAP representation across all adenoma cells. (c) Spatial distribution of colocalised adenoma epithelial cells (upper) and Tregs (lower). (d) Ligand activity between colocalised single cells from epithelial adenoma cells colocalised with Tregs to other cells. The widths of the lines correspond to the ligand activity scores. (e) Imputed MDK expression in spatial distribution. (f) MDK normalised expression in colocalised cell populations in UMAP representation. (g) Violin plot representing MDK normalised expression in colocalised adenoma epithelial cells and other epithelial adenoma cells. ∗∗∗p < 0.001. p values were determined using the Wilcoxon rank-sum test and Benjamini–Hochberg method. (h) Volcano plot representing the differentially expressed ligand genes between colocalised adenoma epithelial cells and other adenoma epithelial cells. p values were determined using the Wilcoxon rank-sum test and Benjamini–Hochberg method. (i) Gene ontology (GO) analysis of biological process comparing colocalised adenoma epithelial cells and other adenoma epithelial cells.