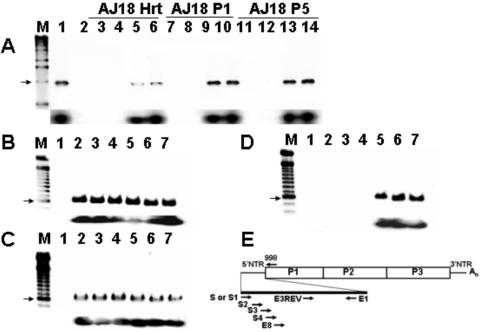
FIG. 3.
Primers at the 5′ end of the viral RNA sequence fail to amplify cDNA from day 18 heart homogenates or heart homogenate passages. (A) RT-PCR of RNA from AJ d18 p.i. heart homogenate (AJ18 Hrt) or from cultures of homogenate passed once (AJ18 P1) and five times (AJ18 P5)t in HeLa cells. cDNA was amplified using different 5′ primers and a constant downstream primer, 998. Lane M, 1-kb marker (the arrow shows a 1-kb band); lane 1, RT-PCR of CVB3/28 RNA with S-998 amplimer (1,021 bp); lane 3, negative control RT-PCR; lanes 3, 7, and 11, primer S; lanes 4, 8, and 12, primer S2; lanes 5, 9, and 13, primer S3; lanes 6, 10, and 14, primer S4. Southern blot analysis (lower panel) with primer E3REV confirms that amplified cDNA is CVB3. (B-D) RT-PCR analysis shows that 5′ termini can be amplified from 105 HeLa cells infected with parental CVB3/28 (B) and AJ14 P5 (RNA heart homogenate from a mouse killed 14 days p.i. and passed five times in HeLa cells) (C) but not from AJ18 P5 (D). Bottom portions of panels B to D show Southern blots of gel probed with E3REV. Lanes M, 100-bp markers (each arrow shows a 600-bp band). Lane 1, negative control; lanes 2 to 7, infected cell RNA. Different 5′ end primers were assayed with the downstream primer E1. Lanes 1 and 2, 5′ primer is S; lane 3, 5′ primer is S1; lane 4, 5′ primer is S2; lane 5, 5′ primer is S3; lane 6, 5′ primer is S4; lane 7, 5′ primer is E8 (Table 1). (E) Relative positions in the CVB3 genome of primers used.