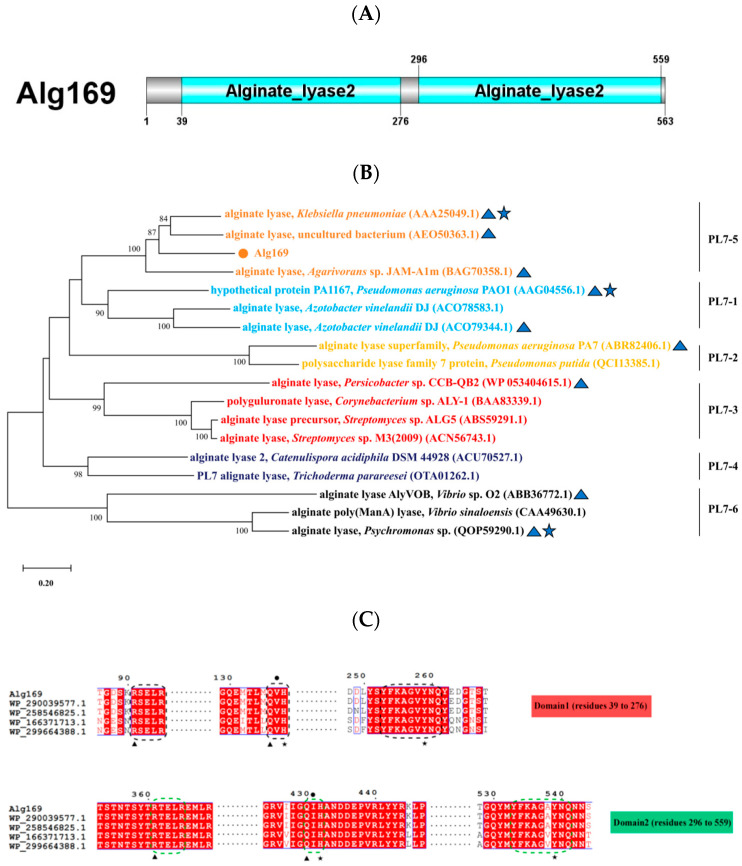
Figure 2.
(A) Schematic representation of Alg169 domain configuration. (B) The phylogenetic tree of Alg169 and other alginate lyases within the PL7 family was constructed using MEGA X. Bootstrap values for each branch were obtained through 1000 repetitions. Alg169 is indicated by a spot, the characterized alginate lyases are shown by triangles, and known structure alginate lyases are marked with pentagrams. (C) Result of alignment analysis of Alg169 and related alginate lyases (WP_290039577.1, alginate lyase of Psychromonas sp. 14N.309.X.WAT.B.A12; WP_258546825.1, alginate lyase of Psychromonas sp. B3M02; WP_166371713.1, alginate lyase of Psychromonas sp. SA13A; and WP_299664388.1, alginate lyase of uncultured Psychromonas sp.). The black and green dashed boxes indicate the conserved motifs for the catalytic activity of the PL7 family in Domain 1 and Domain 2, respectively. Neutralizing residues and catalytic acids/bases are marked with black triangles and stars. Black circles pinpoint amino acids related to catalytic activity. Amino acids that are conserved across the sequences are highlighted with a red background and white character.