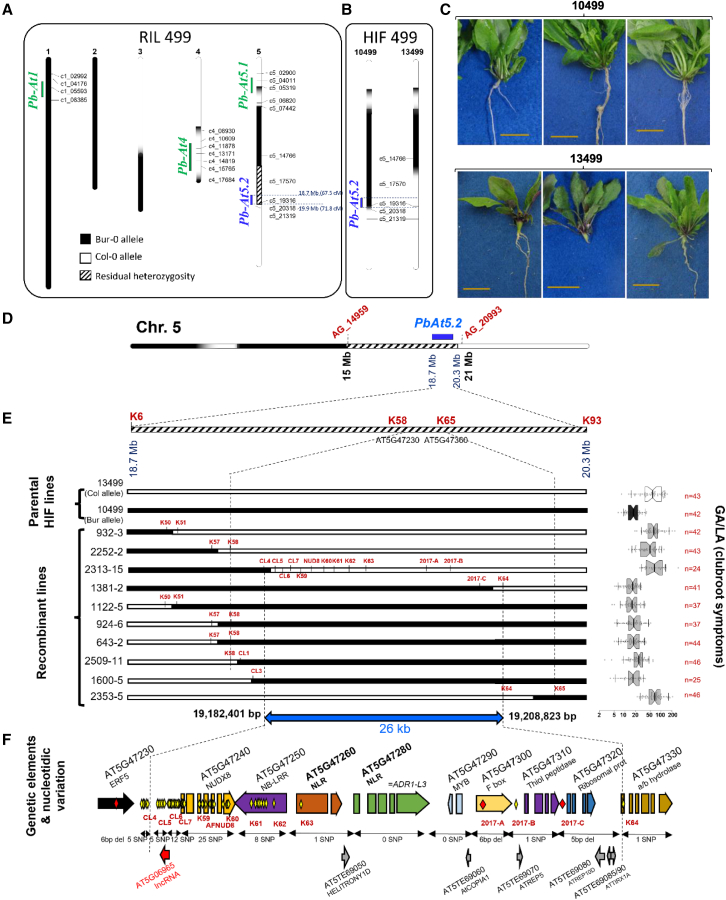
Figure 1.
Fine mapping of Pb-At5.2.
(A) Genetic map and residual heterozygosity in the recombinant inbred line (RIL) 499 derived from Bur-0 and Col-0 (Alix et al., 2007), and genetic and physical positions of clubroot resistance QTLs (Jubault et al., 2008b). Black, Bur-0 allele; white, Col-0 allele; hatched, heterozygous (Col-0/Bur-0).
(B) Allele configuration at Pb-At5.2 in the two derived HIF lines 10499 and 13499.
(C) Photos showing that Pb-At5.2 conferred partial resistance to the eH isolate in the HIF 499 genetic background. HIF 10499 and 13499 harbored Bur-0 and Col-0 alleles, respectively, at Pb-At5.2. Observations were made 21 days post inoculation.
(D) First round of fine mapping: allelic structure in the F1 lines derived from reciprocal crosses between 10499 and 13499. A total of 554 F3 lines with recombination in the confidence interval were screened using 10 SNP markers between AG_14959 and AG_20993. High-density genotyping (94 SNPs from K1 to K94) in 88 recombinant F3 lines and clubroot phenotyping of their bulked segregating F4 progenies led to the identification of a new interval between markers K58 (AT5G47230) and K65 (AT5G47360).
(E) Second round of fine mapping: recombination positions in homozygous individuals obtained from selected recombinant lines. For each line, the GA/LA index (disease symptoms, log scale) is indicated on the right panel. Center lines show the medians; box limits indicate the 25th and 75th percentiles as determined by R software; whiskers extend 1.5 times the interquartile range (IQR) from the 25th and 75th percentiles; outliers are represented by dots; data points are plotted as open circles. The number of individual plants analyzed for each genotype is indicated (n). The notches are defined as +/− 1.58 × IQR/sqrt(n) and represent the 95% confidence interval for each median. GA/LA values statistically different from those of 13499 are indicated by asterisks (t-test, ∗∗∗p < 0.001). Genetic markers are indicated for each recombination position. Markers between CL5 and 2017-C were used in every line but are shown only for 2313-15.
(F) New 26-kb interval of Pb-At5.2 between markers CL4 (excluded) and K64 (excluded), containing eight annotated ORFs, six transposons, and one long non-coding RNA. Yellow and red diamonds indicate SNPs and nucleotide deletions, respectively.