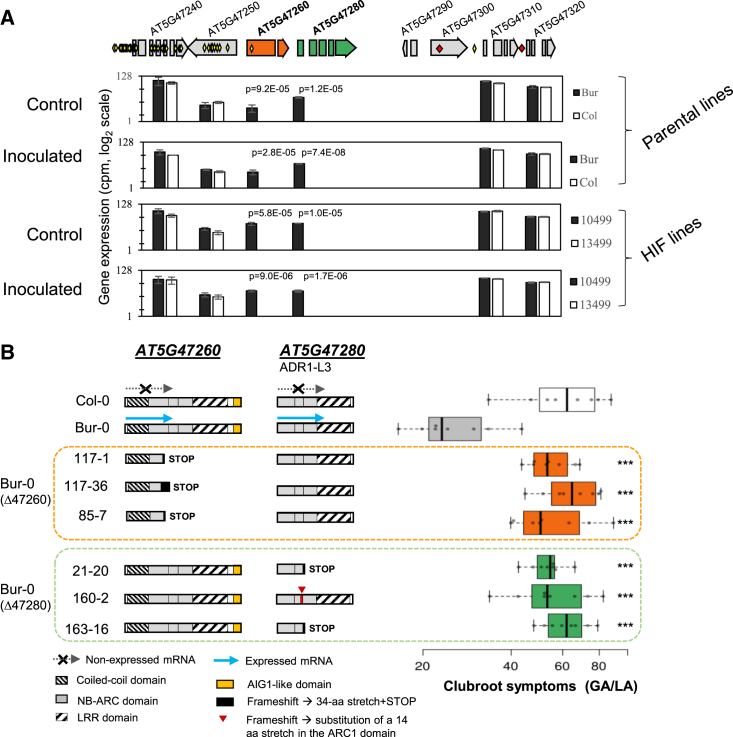
Figure 2.
Identification and CRISPR-Cas9 validation of two NLR-encoding genes controlling clubroot resistance at QTL Pb-At5.2.
(A) Sequence variations and expression levels of genes in the Pb-At5.2 region. Gene expression values are from RNA-seq analyses conducted under inoculated and control conditions at 14 days post inoculation (log2 normalized CPM) with parental lines Col-0/Bur-0 and HIF lines 13499/10499 (the last two were derived from RIL 499 and are homozygous Pb-At5.2Col/Col and Pb-At5.2Bur/Bur, respectively) (Supplemental Data 2). False discovery rate–adjusted p values are shown if less than 0.05. Yellow and red diamonds indicate SNP and INDEL variations, respectively.
(B) Effect of AT5G47260 or AT5G47280 knockout on GA/LA index (disease symptoms) in the Bur-0 background. Cas9-mediated mutations were obtained in the Bur-0 genetic background. For each targeted gene, three independent lines harboring independent homozygous mutations were used. Lines 117-1 and 21-20 no longer have the CRISPR-Cas9 cassette. For each line, the mean clubroot symptom score (GA/LA, log scale) was obtained by modeling raw data of eight biological replicates (with 10–12 individual plants per replicate). Center lines show the medians; box limits indicate the 25th and 75th percentiles as determined by R software; whiskers extend 1.5 times the IQR from the 25th and 75th percentiles; data points are plotted as open circles. GA/LA values of edited lines that are statistically different from the Bur-0 GA/LA value are indicated by asterisks (Dunnett’s test) as follows: ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.