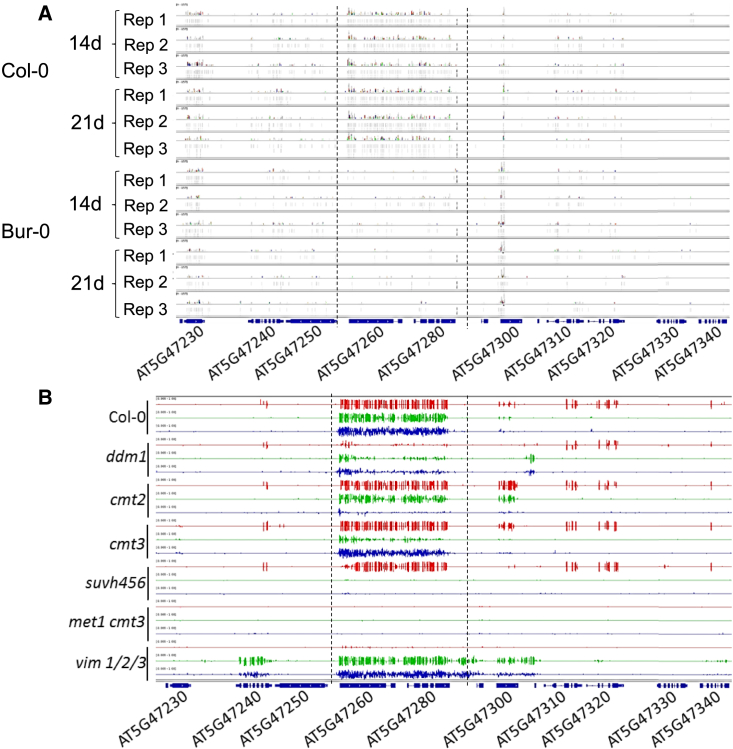
Figure 7.
Epigenetic variation at Pb-At5.2 correlates with the abundance of locus-targeted sRNA but is maintained by the RNA-independent methylation pathway.
(A) Mapping of sRNA-seq reads. Reads were obtained from roots of Col-0 and Bur-0 accessions at two time points 14 and 21 days after sowing. For each condition, three bulks (numbered from Rep 1 to Rep 3) of six plants were used.
(B) Methylation state at the Pb-At5.2 locus in knockout lines (Col-0 genomic background) defective for the RdDM or non-RdDM pathway (Stroud et al., 2013).
Red: methylation in CG context. Green: methylation in CHG context. Blue: methylation in CHH context.