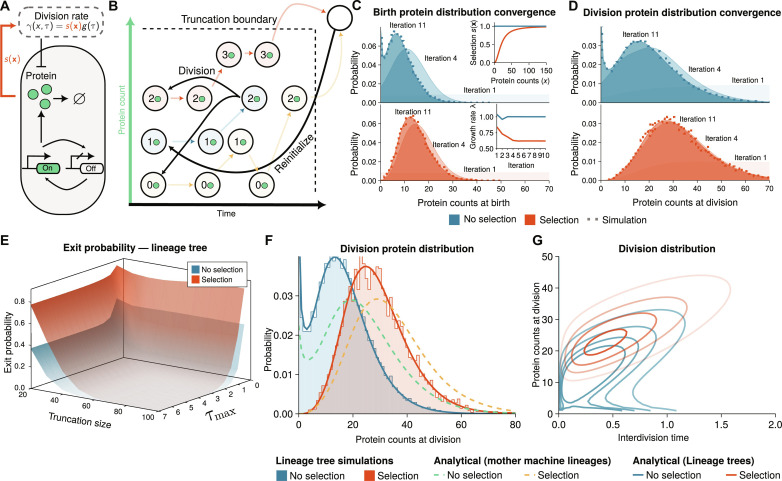
Fig. 2. FSP enables accurate prediction of selection on gene expression noise.
(A) Schematic illustration of the agent-based model. Intracellular dynamics of stochastic gene expression are modeled by the telegraph model for bursty expression of a protein. The protein is binomially partitioned at cell division. The selection effect is introduced through a division rate increasing with expression (details in section S3A). (B) Illustration of the finite state projection (FSP) algorithm. Intuitively, gene expression (light arrows) drives cells to either commit to division or cross the truncation boundary, leading to their states being reinitialized (dark arrows). (C and D) Birth and division distributions for the model converge in a few iterations and agree with agent-based simulation of lineage trees obtained using the first division algorithm (Materials and Methods; see fig. S1 for mother machine lineages). Top inset of (C) shows the used selection functions [s(x) = 1 for no selection, s(x) = 1/((20/x)2 + 1) for selection on the gene expression]. Bottom inset shows the convergence of the computed population growth rate λ (see Materials and Methods). (E) The exit probability due to FSP truncation decreases with truncation size and time horizon τmax. (F) Summary distributions without (blue solid line) and with selection (red solid line) are compared to direct simulations of the lineage trees (shaded areas). Our method is in agreement with the simulation results. Distributions obtained using mother machine lineages (dashed) are also shown. (G) FSP solution of the agent-based model predicts bimodal division distribution without division-rate selection (blue lines correspond to levels of equal probability) and an unimodal division distribution in the presence of division-rate selection.