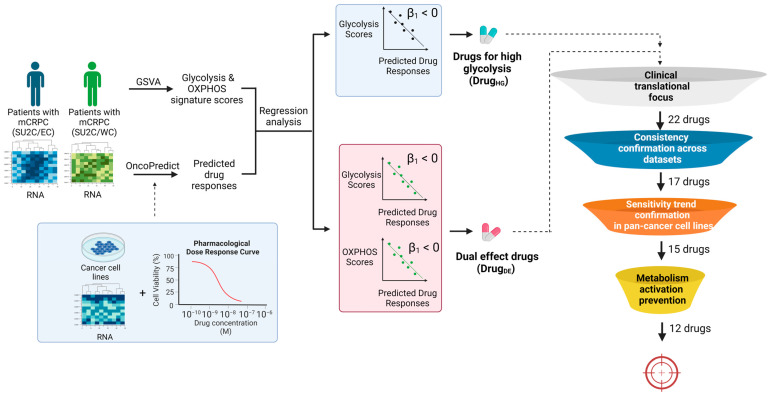
Figure 1.
Lead compound nomination pipeline for mCRPC patients with high glycolysis activity. OncoPredict was used to impute sensitivities of mCRPC patient tumors to various drugs. GSVA was used to compute glycolysis and OXPHOS scores for each mCRPC patient tumor. Subsequently, regression analysis was conducted. Drugs with predicted drug response significantly and negatively correlated with glycolysis scores or both glycolysis and OXPHOS scores were selected as drugs for high glycolysis (DrugHG) and dual-effect drugs (DrugDE), respectively. Only drugs identified in both SU2C/EC and SU2C/WC studies were included for subsequent filtering steps to select final drug candidates. The four filtering steps included (1) clinical translational focus, (2) consistency confirmation across datasets, (3) sensitivity trend confirmation in pan-cancer cancer cell lines, and (4) metabolism activation prevention. Abbreviations: mCRPC: metastatic castration-resistant prostate cancer; SU2C/EC: Standard Up to Cancer East Coast; SU2C/WC: Standard Up to Cancer West Coast; GSVA: Gene Set Variation Analysis; β1: a coefficient of predicted drug response of mCRPC tumors; OXPHOS: oxidative phosphorylation.