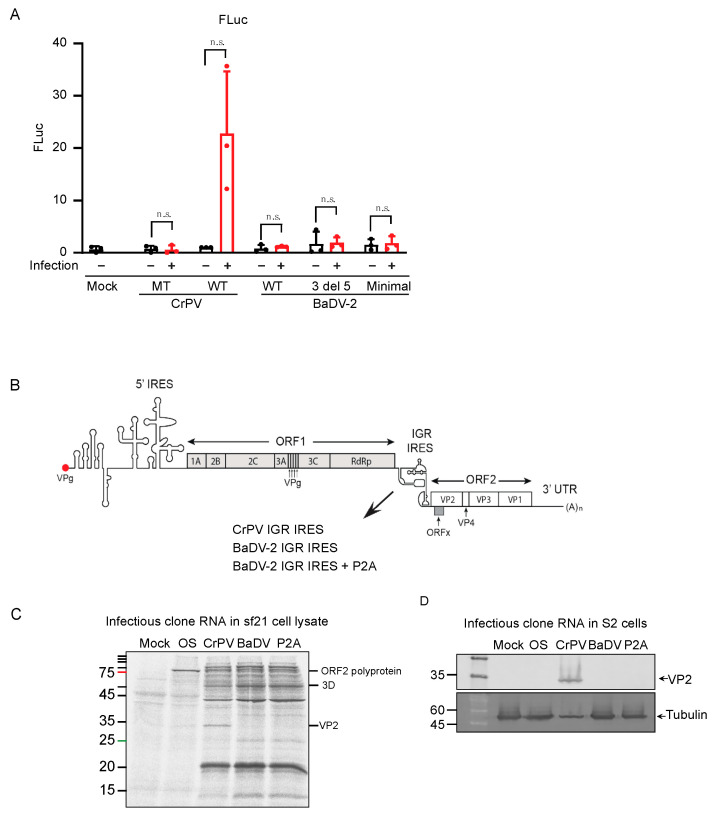
Figure 6.
BaDV-2 IGR IRES activity in infectious clone and in cells. (A) Bicistronic reporters were transfected into S2 cells followed by mock infection or CrPV infection (MOI = 20). Cells were collected 6 h post transfection. Bicistronic RNAs tested contain the wild-type CrPV, mutant CrPV (CC6214-5 to GG to disrupt PKI base-pairing), wild-type BaDV-2 full-length IGR IRES (WT BaDV-2), BaDV-2 3′ DEL 5, and BaDV “minimal” IRES (double deletion at both 3′ and 5′ end. Luciferase activities were measured and normalized to wild-type CrPV with mock infection. Red and black dots represent data points from three independent experiments. A paired t-test was used to determine the p value and thus the significance levels. “n.s.” denotes the difference is not significant between the control groups and the experimental groups (p > 0.05). Shown are the averages from at least three independent experiments ± standard deviation. (B) Schematic of infectious clone CrPV (CrPV, CrPV_IGR_IRES) and chimeric clones with CrPV IGR IRES replaced with BaDV-2 IGR IRES (BaDV-2 IGR IRES) and with P2A-site added after BaDV-2 IGR IRES (BaDV-2 IGR IRES+ P2A). (C) Infectious clone RNAs were incubated in Sf-21 extracts containing [35S]-methionine/cysteine and then monitored by SDS-PAGE analysis. Mock = mock transfection; OS = ORF1Stop; CrPV = pCrPV; BaDV = BaDV-2 IGR IRES; P2A = BaDV-2 IGR IRES + P2A. Shown is a representative gel from at least three independent experiments. (D) In vitro transcribed infectious clone RNAs were transfected into S2 cells for 144 h. VP2 expression was detected by immunoblotting. Shown are a representative SDS PAGE gel and the averages from at least three independent experiments ± standard deviation.