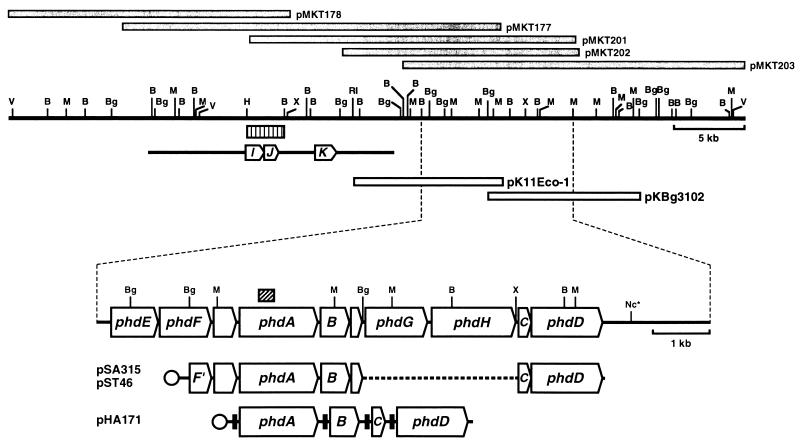
FIG. 1.
Physical map and genetic organization of phenanthrene-degrading genes. Gray bars indicate the DNA inserts in the library clones. Open bars show inserts in the two clones used in this study. Arrows on the enlarged physical map show the locations and orientations of the phenanthrene-degrading genes: phdA, the α subunit of the oxygenase component; B, phdB, the β subunit of the oxygenase component; C, phdC, the ferredoxin component; phdD, the ferredoxin reductase component; phdE, dihydrodiol dehydrogenase; phdF, extradiol dioxygenase; phdG, hydratase-aldolase; phdH, aldehyde dehydrogenase. The box with oblique indicates the 302-bp PCR-amplified fragment that was used for probing the library clones. The typical expression plasmids are shown at the bottom. Circles indicate promoters. Transcription of DNA segments containing phdABCD genes cloned in pSA315 and pST46 is under the control of the lac and T7 promoters, respectively. The transcription in pHA171 of the phdABCD genes preceded by the SD sequence that is effective in E. coli (filled boxes) is under the control of the T7 promoter. The bar with three arrows indicates the previously reported sequence (AB000735 in DDBJ/GenBank/EMBL databases), arrows marked with I, J, and K showing the locations and orientations of the phenanthrene-degrading genes phdI (1-hydroxy-2-naphthoate dioxygenase), phdJ (trans-2′-carboxybenzalpyruvate hydratase-aldolase), and phdK (2-carboxybenzaldehyde dehydrogenase). The striped bar shows the 2.5-kb HindIII-BamHI DNA fragment that was used as probe in the hybridization following PFGE (Fig. 2). Restriction sites: B, BamHI; Bg, BglII; H, HindIII, M, MluI; Nc, NcoI; RI, EcoRI; V, EcoRV; X, XbaI. The asterisk indicates that the other NcoI restriction sites are not shown.