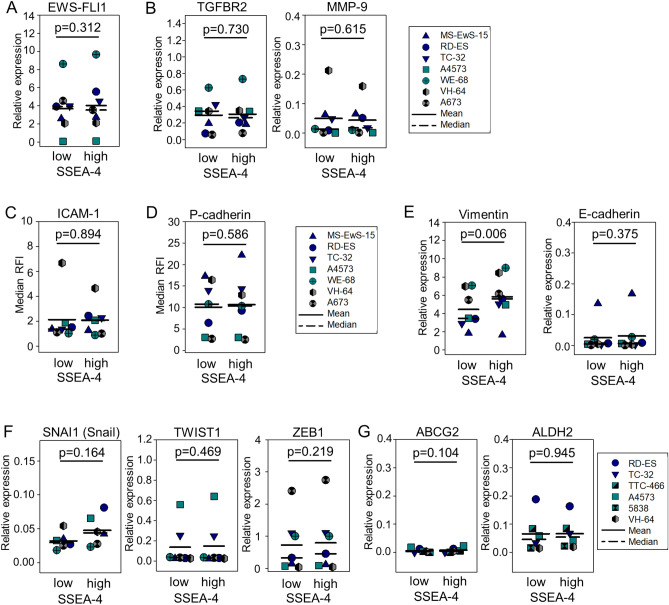
Figure 3.
SSEA-4high phenotype is not associated with expression of EWSR-FLI1 and with epithelial/mesenchymal plasticity. (A) Expression of the EWSR1-FLI-1 fusion transcript in SSEA-4low and SSEA-4high sorted subpopulations of 7 EwS cell lines was quantified by qRT-PCR. Triplicates of mRNA isolated from 2–3 independent sorting experiments per cell line were analyzed. Relative expression was determined by calculating Delta Ct with a reference gene. (B) Gene expression of TGFBR2 and MMP-9 by qRT-PCR analysis as above. (C) ICAM-1 and (D) P-cadherin surface expression in 7 EwS cell lines by flow cytometry using APC-labeled antibodies against ICAM-1 or anti-P-cadherin, respectively. Costaining with anti-SSEA-4 PE antibody allowed to compare ICAM-1 expression levels between SSEA-4low versus SSEA-4high expressing cells by gating on the 10% lowest and 10% highest SSEA-4 expressing cells. (E) Expression of genes encoding Vimentin and E-Cadherin, (F) EMT transcriptions factors SNAI1 (Snail), TWIST1 and ZEB1, and (G) markers associated with cancer-initiating cells (ABCG2, ALDH2) in SSEA-4low and SSEA-4high subpopulations of 6 or 7 EwS cell lines (TC-32 not analyzed for SNAI1) were quantified by qRT-PCR as described above. After testing for normal distribution, statistical significances for E-cadherin, TWIST1 and ZEB1 (data not normally distributed) were determined with the Wilcoxon signed rank test and in all others (normal distribution) with the paired student’s t-test.