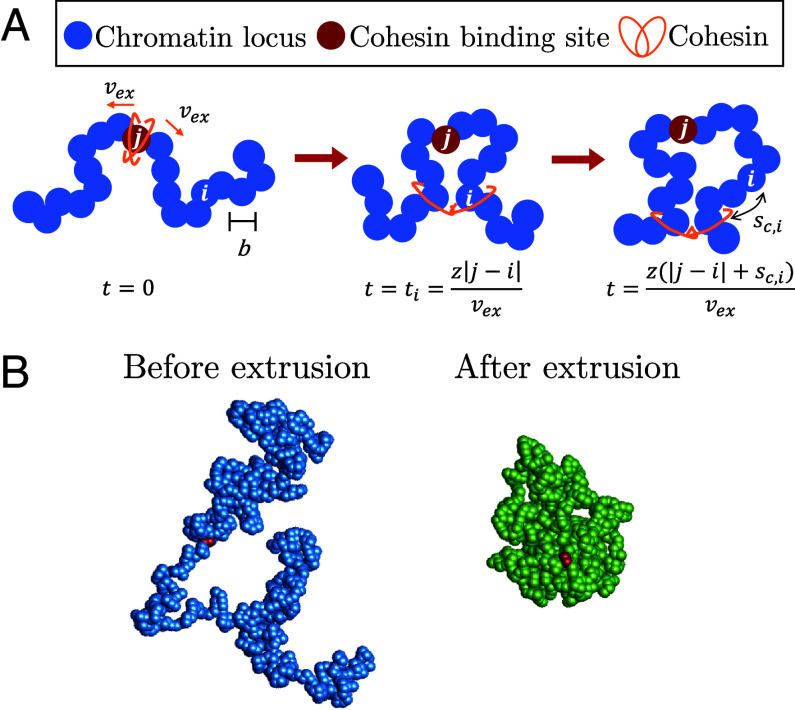
Fig. 4.
Schematic of active loop extrusion model and simulation snapshots. (A) Chromatin is discretized into loci with size b each representing base pairs. Cohesin domains bind to locus j and extrude away from one another with average curvilinear velocity in units of the number of base pairs per unit time. Locus i is extruded at the time . At time , locus i is base pairs away from the cohesin. (B) Snapshots of a hybrid MD—MC simulation of a single active loop extrusion cycle when a single cohesin extrudes a single bead-spring polymer chain once. The snapshots are before extrusion starts with (Left, blue) and after the cohesin reaches the chain ends (Right, green). Each bead represents 1 kbp of chromatin (see SI Appendix for details). The red beads indicate the cohesin binding site.