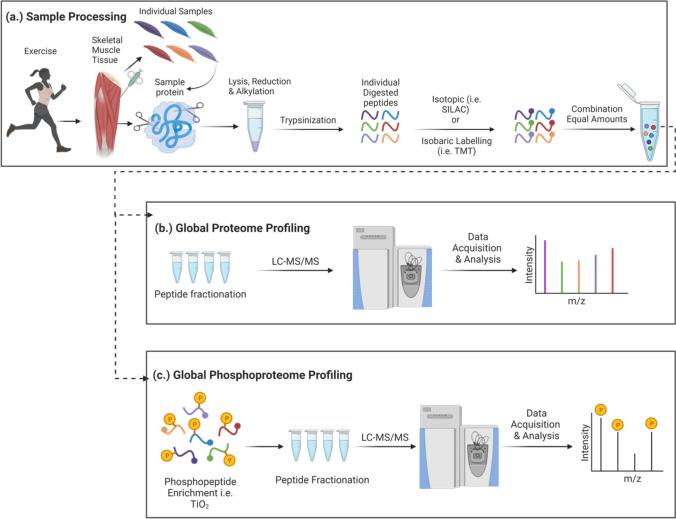
Fig. 4.
Proteomic and phosphoproteomic sample preparation and analysis workflow overview. a In MS-based phosphoproteomic analysis workflows, proteins are first homogenised and extracted from skeletal muscle samples. Following protein extraction, the sample preparation workflow then involves protein extraction, denaturing, reduction and alkylation followed by trypsin digestion into peptides. Isotopic [i.e. stable isotope labelling by amino acids in cell culture (SILAC)] or isobaric labelling [i.e. tandem mass tag (TMT)] of peptides is commonly performed to improve the sensitivity and throughput of sample analyses by combining tagged samples, whereby labelled peptides from each sample are combined in equal amounts and can be distinguished from each other by MS; b this combined labelled sample can then be further fractionated to reduce the overall sample complexity; and c in global phosphoproteome profiling, phosphopeptides are enriched [e.g. using titanium dioxide (TiO2)] and fractionated with liquid chromatography (LC) before being identified and quantified by LC–MS/MS analysis. Finally, total peptides and/or phosphopeptides are identified by their mass to charge ratios (m/z) and phosphorylation site positions are assigned [178]. Adapted from ‘global protein and phospho profiling’, by BioRender.com (2023). Retrieved from https://app.biorender.com/biorender-templates