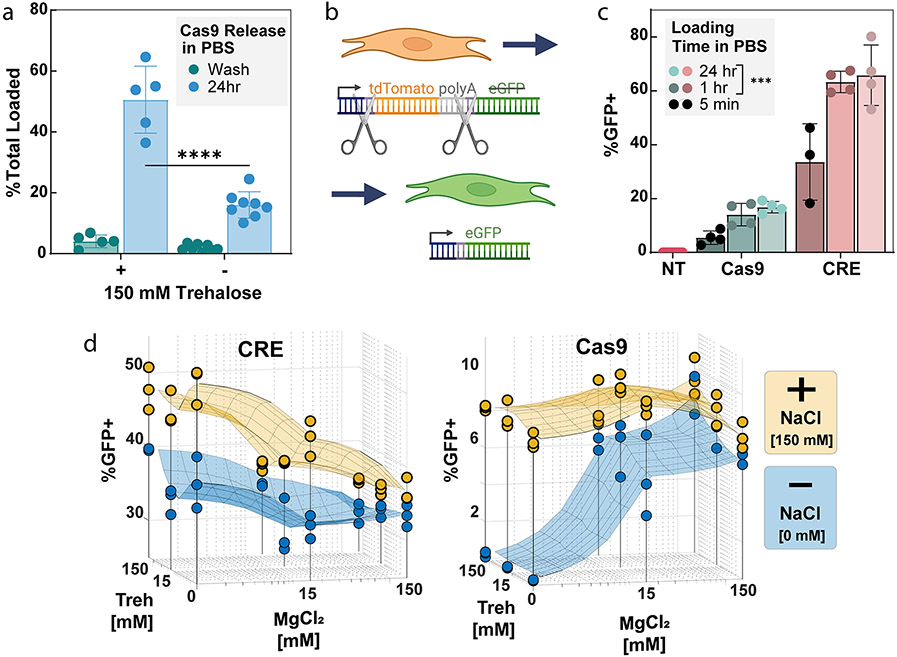
Figure 2.
Effects of excipient and salts in loading buffer and loading time on editing activity. (a) Cargo loading is stable based on negligible release during a wash, spin-down, and resuspension of the PSiNPs, with or without addition of 150 mM trehalose in the loading buffer. However, addition of trehalose during Cas9 loading increases release of Cas9 RNP from the PSiNPs over 24 h (****p ≤ 0.0001, Šídák’s multiple comparisons test). Each point on the graph represents an independent replicate. (b) Schematic of the mTmG reporter NIH 3T3 fibroblasts. The mTmG reporter cassette was inserted into the Rosa26 genomic locus. Cas9 or CRE recombinase targets LoxP sites within the cassette. Successful editing removes tdTomato and the polyadenylation tail, turning on eGFP expression. (c) Effect of time of cargo incubation during the PSiNP loading effect on gene editing activity. NT = No Treatment, statistical comparison vs 5 min loading time group, ***p ≤ 0.001, Tukey’s multiple comparisons test. (d) Effect of NaCl, MgCl2, and trehalose concentration in loading buffer on positively charged CRE or negatively charged RNP editing activity in cells. In sections c and d, each point represents a separate treatment well, with percent positive measured by flow cytometry. All error bars are standard deviation.