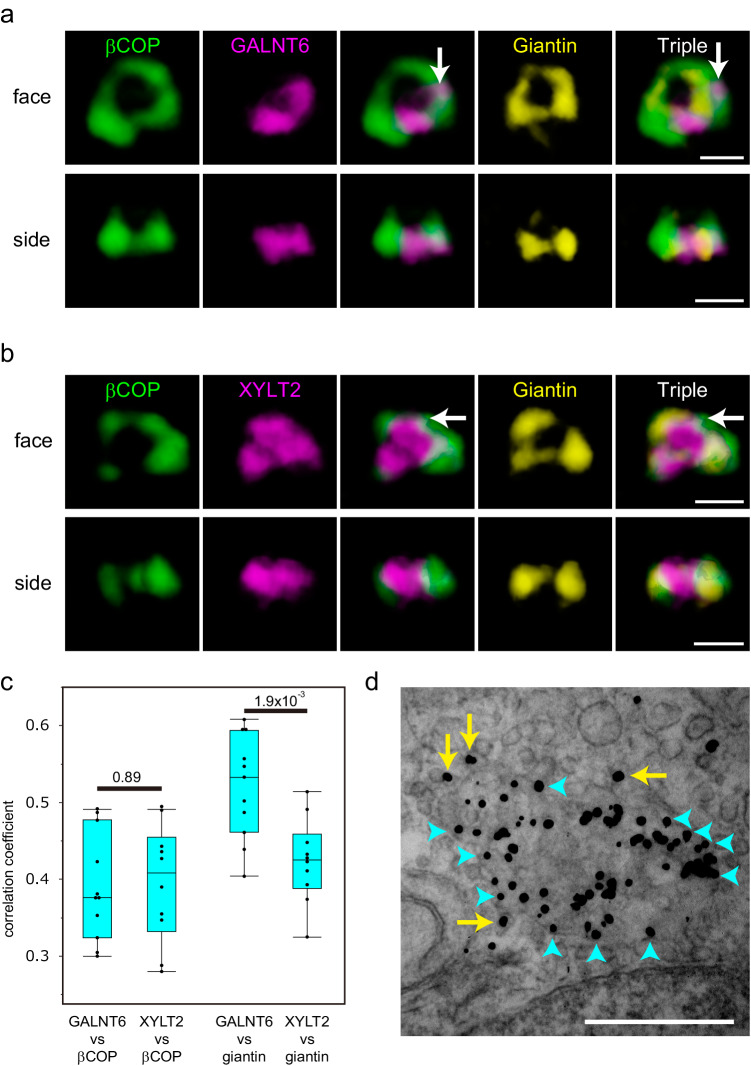
Fig. 3. Distribution of glycosylation enzymes with COPI.
a, b Three-colour SCLIM 3D imaging of isolated Golgi units in KI cells stained with antibodies against βCOP, giantin, and PA tag. The upper two lines of panels show the Golgi in a GALNT6-3xPA KI cell. The lower two lines of panels show the Golgi in a XYLT2-3xPA KI cell. face: en face view, side: side view. The same Golgi unit is observed from different angles in Fig. 3a and 3b. βCOP signals colocalised with GALNT (Fig. 3a) and XYLT2 (Fig. 3b) outside of giantin are shown by arrows. Bar, 1 µm. Images are representative of at least three independent biological replicates. Imaging modalities and acquisition parameters of SCLIM are described in the materials and methods. c 3D colocalisation analysis between GALNT6/XYLT2 and βCOP (left two lanes) and GALNT6/XYLT2 and giantin (right two lanes) in cells without nocodazole. The number of cells: 10 (left two lanes) and 11 (right two lanes). Statistical significance was determined by two-tailed unpaired t test. Each point represents the value from one cell. P values are depicted in the graph. Boxes represent 25% and 75% quartiles, lines within the box represent the median, and whiskers represent the minimum and maximum values within 1.5x the interquartile range. d Immunoelectron micrographs showing the localisation of giantin (arrows) in the en face view a Golgi cisterna. Signals at the edge of the cisterna are shown in blue arrowheads. Signals on the vesicles are shown by yellow arrows. The image is a representative of more than ten cells from three independent experiments. Bars, 500 nm.