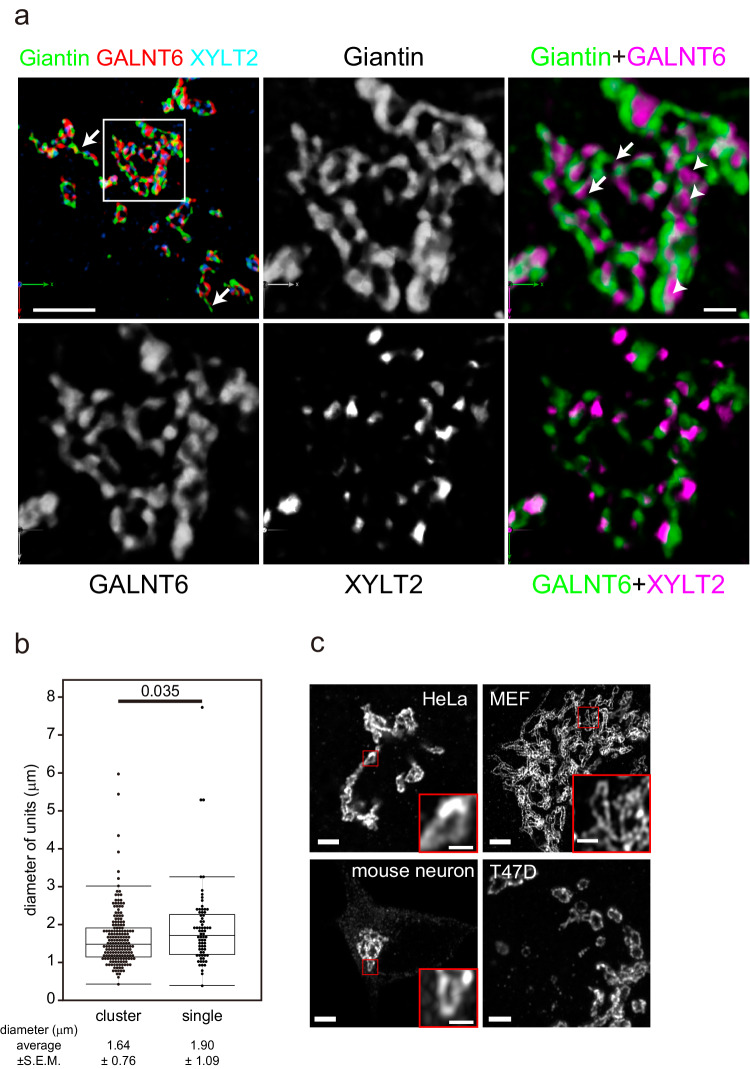
Fig. 4. Architecture of the Golgi in the absence of nocodazole.
a Three-colour SCLIM 3D imaging of fixed GALNT6-9xMyc + XYLT2-3xPA double KI cells. The left upper panel shows the Golgi at low magnification (Supplementary Movie 1). Other panels are magnified views of a rectangle in the left upper panel. The Golgi appears to be a cluster of Golgi units connected directly or by tubes (arrows) positive for giantin. Glycosylation enzymes are largely colocalised with giantin, whereas some enzymes are localised within the units (arrowheads). Bars, 5 µm (left upper panel), 1 µm (right upper panel) b Distribution of the long diameter of the Golgi units in cluster and isolated units by Airyscan. One unit is defined as circular to elongated area encircled by giantin which contains glycosylation enzymes. The number of cells: 6. The number of units: >100 (units in clusters), >70 (single units). Boxes represent 25% and 75% quartiles, lines within the box represent the median, and whiskers represent the minimum and maximum values within 1.5x the interquartile range. Statistical significance was determined by two-tailed unpaired t test. P values are depicted in the graph. c Golgi units whose rims are marked by giantin in different types of cells. Small red squares are magnified as insets. Bars, 2 µm (large panels), 0.5 µm (insets). Images are representative of at least three independent biological replicates. Imaging modalities and acquisition parameters of SCLIM are described in the materials and methods.