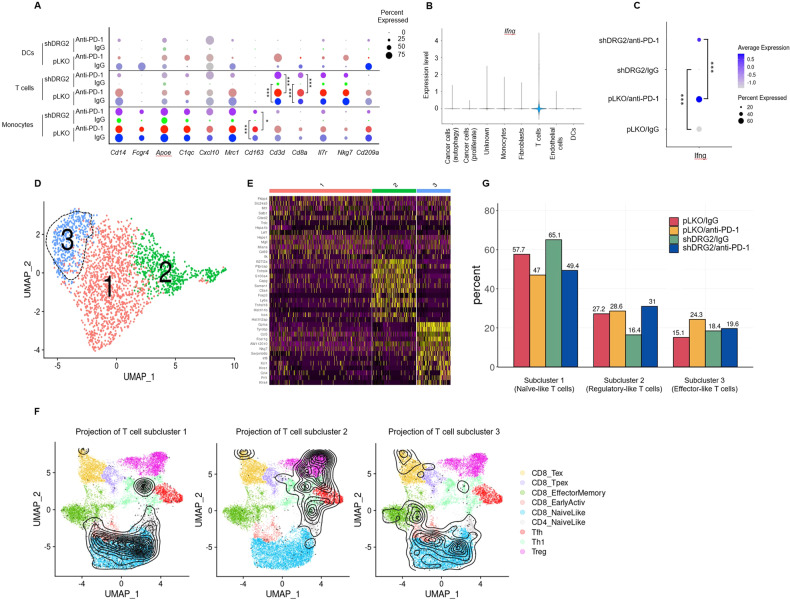
Fig. 7. Anti-PD-1 immunotherapy fails to activate T cells in DRG2-depleted melanoma tumors.
A A dot plot showing the percentage of cells expressing immune-related marker genes across the immune cell clusters in B16F10/pLKO or B16F10/shDRG2 tumors treated with negative control (IgG) or anti-PD-1. Student’s t test. ***P < 0.001. B A violin plot showing the log10 expression of IFN-γ gene across 8 clusters. C A dot plot showing both the average expression IFN-γ and the percentage of cells expressing IFN-γ in B16F10/pLKO or B16F10/shDRG2 tumors treated with negative control (IgG) or anti-PD-1. A darker color indicates higher average gene expression from the cells in which the gene was detected, and larger dot diameter indicates that the gene was detected in greater proportion of cells from the cluster. Student’s t test. ***P < 0.001. D UMAP of 2341 T cells showing 3 subclusters of T cells indicated by the color-coded legend. E A heatmap based on relative expression of the top-10 most differentially expressed genes for each T cell subcluster. F Annotation of T cell subclusters into reference atlas using ProjecTILs. The cells in the colored background refer to the cell types in the reference atlas, and the cells in the black circle represent the cells in the T cell subcluster 1, subcluster 2, and subcluster 3. Subcluster-defining genes are shown in Supplementary Fig. S8. G Relative contribution of each T cell subcluster (%) in B16F10/pLKO or B16F10/shDRG2 tumors treated with negative control (IgG) or anti-PD-1.