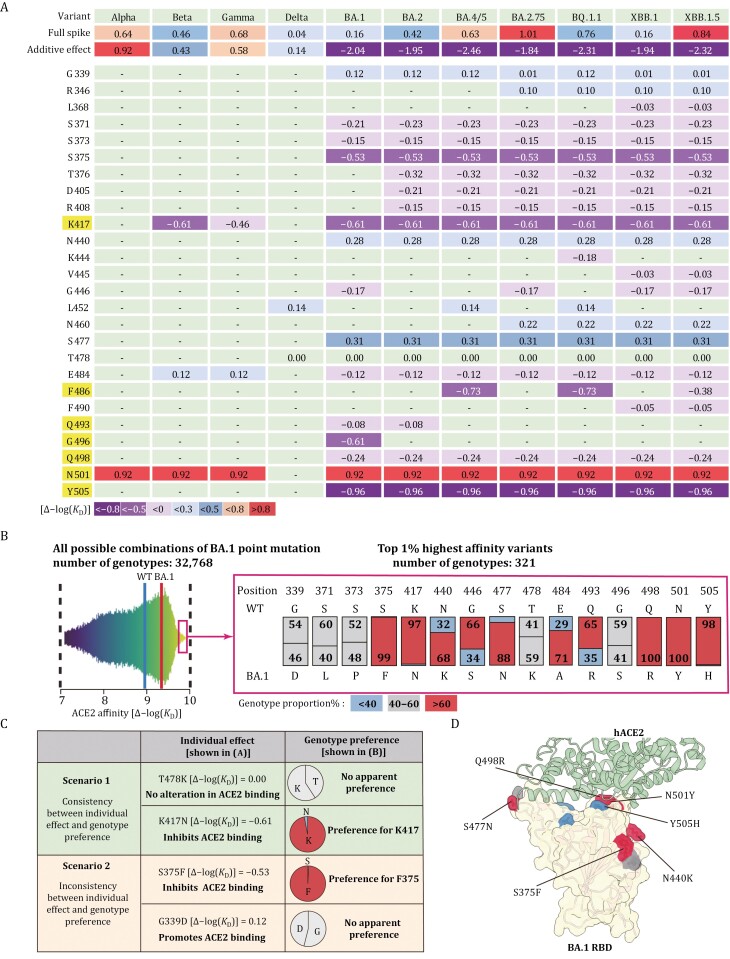
Figure 3.
Potent epistatic mutations reverse the deleterious summed effect on ACE2 affinity. (A) Heat map depicting relative enhancement in ACE2 affinity for variants of concern (VOCs) (top row) and individual mutations (other rows). Each column shows data for each variant, including Alpha, Beta, Gamma, Delta, BA.1, BA.2, BA.4/5, BA.2.75, BQ.1.1, XBB.1, and XBB.1.5. The data in each row are the calculated logarithmic negative KD [Δ−log(KD)] value, to present the effect of mutations on ACE2 binding affinity. These values are calculated by comparing the KD value of a mutated S protein with that of a wildtype S protein under the same experimental condition. The effect for each constituent VOC mutation individually on a wild type background, together with their summed effect is shown. At the left-most column of the table, the original amino acids of the S protein and their corresponding positions are shown; and the amino acids (K417, F486, Q493, G496, Q498, N501, and Y505) directly contacting ACE2 are highlighted. Different colors represent the changed levels on ACE2 affinity: strong enhancement [Δ−log(KD) > 0.8]; moderate enhancement [Δ−log(KD) = 0.5–0.8]; mild enhancement [Δ−log(KD) = 0.3–0.5]; slightly enhancement [Δ−log(KD) < 0.3]; slightly decreased affinity [Δ−log(KD) = −0.5–0]; moderate decreased affinity [Δ−log(KD) = −0.5–−0.8]; strongly decreased affinity [Δ−log(KD) < −0.8]. “−” indicates no mutation at this position for the corresponding variants/column.(B) Systematic analysis of ACE2 binding affinity for RBD proteins containing all possible combinations of 15 mutations in the RBD of BA.1 variant. Left: distribution of ACE2 binding affinity using all possible mutational intermediates of BA.1 (N = 215 = 32,768 RBD genotypes tested). Binding affinity is shown as −log(KD). The vertical lines indicate the −log(KD) for wildtype Wuhan-Hu-1 strain and Omicron BA.1 variant, respectively. An interactive data browser is available at Github. Right: among the top 1% intermediates with a superior ACE2 affinity (a total of 321 genotypes), relative proportions (%) of each amino acid are shown at all 15 mutation sites. On these RBD positions, the amino acids for BA.1 variant and the corresponding amino acid in wildtype Wuhan-Hu-1 strain are also shown. Colors depict the genotypes preference at each position: preferred (genotype proportion > 60%); no apparent preference (genotype proportion = 40%–60%); unappreciated (genotype proportion < 40%). (C) Two scenarios when comparing the effect of individual BA.1 constituent mutations in the wildtype backbone [Δ−log(KD) values from Fig. 3A] with their proportions among the top 1% variants with highest ACE2 affinity (genotype proportions from Fig. 3B). (D) Co-crystal structure of Omicron BA.1 RBD and ACE2 receptor (PDB ID 7WPB). Mutated residues are shown, and their surfaces are colored as the corresponding residues in Fig. 3B.