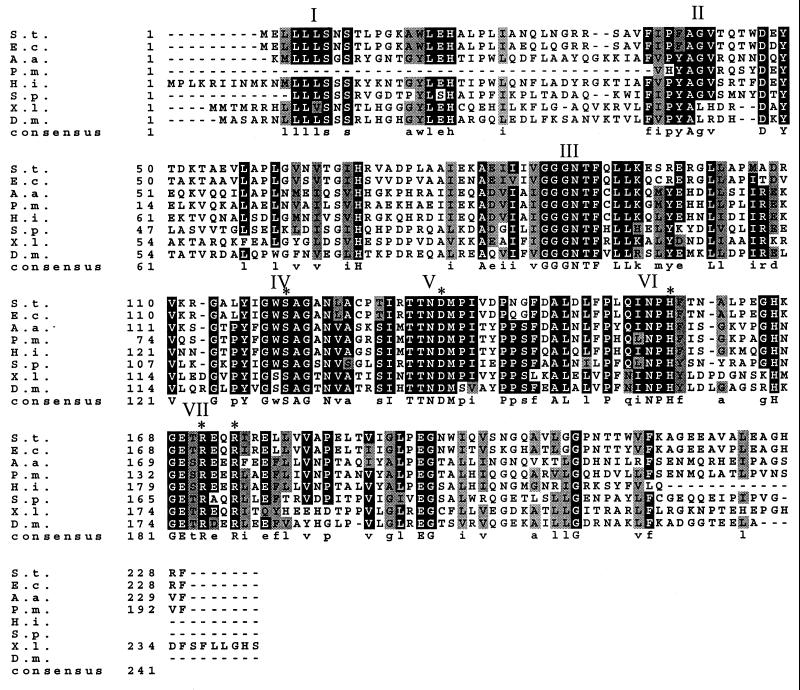
FIG. 1.
Multiple alignment of the amino acid sequences of the peptidase E family. The sequence alignment was determined with ClustalW, and Boxshade 3.1 was used to shade positions at which 70% of the sequences are identical (black) or similar (gray). The residues that are essential for enzyme activity (Ser120, Asp135, His157, Arg171, and Arg174) are indicated with asterisks. Blocks of conserved residues are labeled with roman numerals for clarity when mentioned in the text. Abbreviations are as follows: S.t., S. enterica serovar Typhimurium; E.c., E. coli; A.a., A. actinomycetemcomitans; P.m., partial sequence from P. multocida; H.i., H. influenzae; S.p., S. putrefaciens; X.l., X. laevis; and D.m., D. melanogaster.