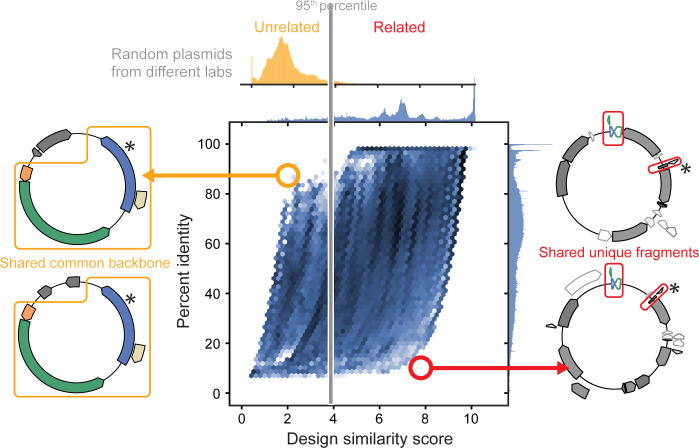
Fig 4. Design similarity scores reliably identify plasmids that are likely to be related while percent identity does not.
The distributions of DS scores and percent identities for pairwise comparisons of plasmids that share undocumented part variants are plotted. Every plasmid containing a given genetic part variant that was observed 1205 or fewer total times was compared to every other plasmid with that part variant for a total of 7,508,114 comparisons. High pairwise percent identity is not compelling evidence that plasmids are related when they share a commonly used backbone, as illustrated by the plasmid pair shown to the left. The DS score of these two plasmids is low in this instance. Low pairwise percent identity also does not necessarily indicate that plasmids are unrelated, as illustrated by the plasmid pair shown to the right. In this case, a high DS score highlights small, but unique sequences present in both plasmids, which is evidence of shared authorship. Asterisks indicate the location of the shared mutation in the associated genetic part variant that differentiates it from the canonical sequence in the annotation database. The distribution of DS scores between 100,000 randomly selected pairs of plasmids from different labs is shown above the plot. The grey line indicates the 95th percentile of the distribution, which was used as the score cutoff for shared plasmid authorship.