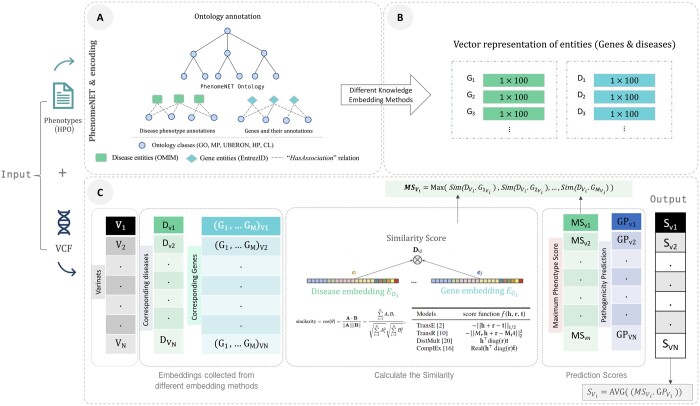
Figure 1.
EmbedPVP Model Workflow. (A) Generates background knowledge from different ontologies. (B) Generates embeddings for diseases (Di) and genes (Gi) using various embedding methods. (C) Calculates phenotype–genotype similarity using the scoring function associated with the selected embedding method, considering the maximum similarity score for multiple genes associated with the phenotype, and then averages this similarity with pathogenicity prediction. Vi represents variant i, MSvi is max phenotype similarity for variants, GPvi is genotype prediction (CADD), and Svi is the final weighted score of phenotypes and genotypes for the variants.