Fig. 2 |. Schematics of the ATR pathway.
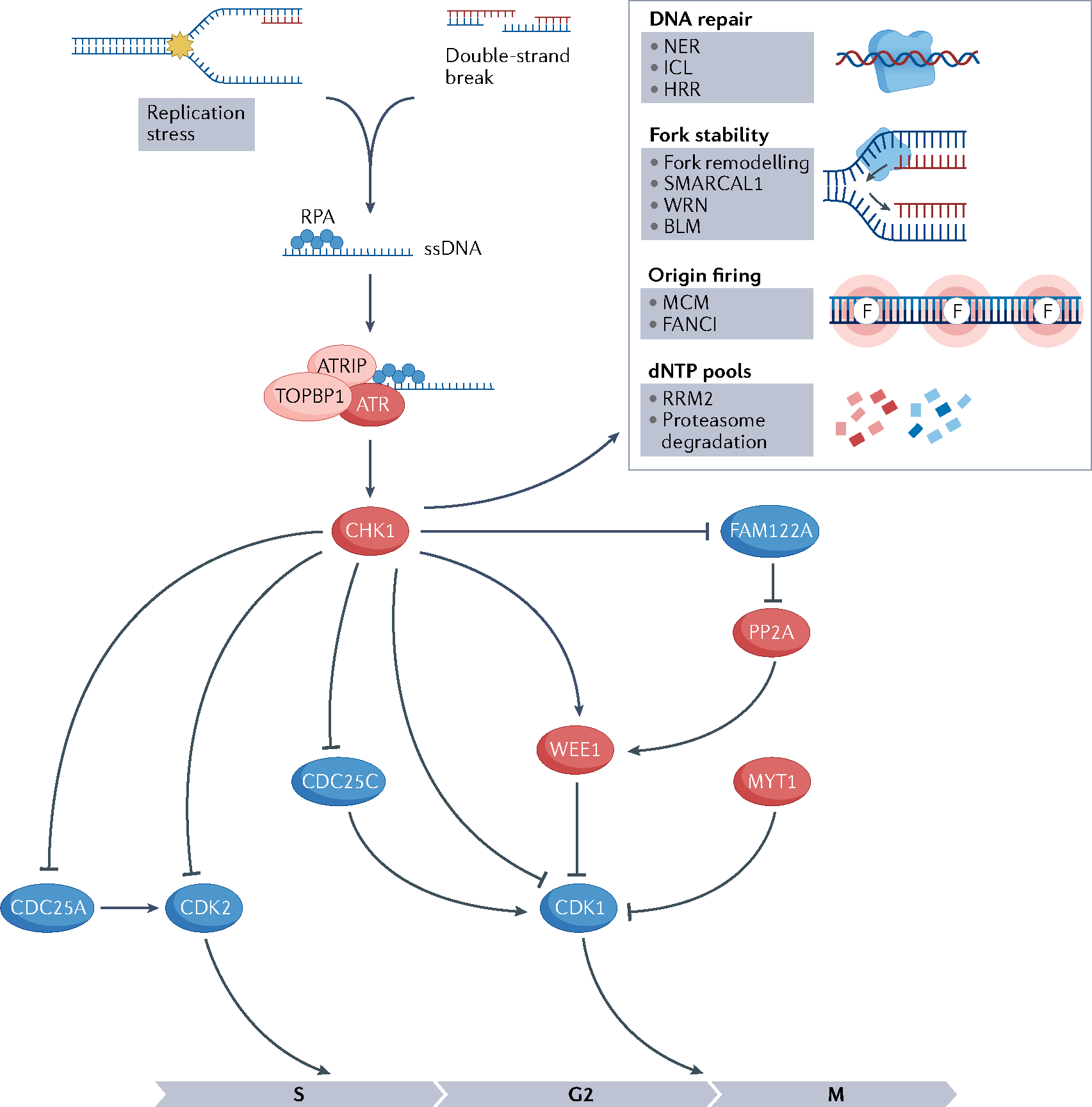
Activation of ataxia telangiectasia mutated (ATM) and Rad3-related (ATR) pathway because of replication stress or double-strand breaks results in cell cycle arrest, activation of DNA repair pathways, fork stabilization, inhibition of origin firing, decrease in deoxynucleotide (dNTP) degradation and increase in dNTP synthesis. Barriers to replication fork progression (shown by the yellow star) lead to the uncoupling of helicases and polymerases, generating single-stranded DNA (ssDNA) at the stalled fork. Replication protein A (RPA) binds to ssDNA at the stalled fork and recruits ATR through the partner ATR-interacting protein (ATRIP). ATR is then activated by DNA topoisomerase 2-binding protein 1 (TOPBP1), activates CHK1 through phosphorylation and triggers the ATR–CHK1 pathway. CHK1 mediates cell cycle arrest in the S–G2 phase through phosphorylation and reduction of cyclin-dependent kinase 2 (CDK2) activation. CDK1 is negatively regulated via phosphorylation by WEE1 and MYT1 kinases and positively regulated by CDC25 phosphatases. CHK1 phosphorylates and inactivates CDC25C, activates WEE1 and promotes the degradation of CDC25A11,13. FAM122A is a negative regulator of the G2/M checkpoint and forms a complex with the phosphatase PP2A. When CHK1 phosphorylates FAM122A, PP2A is disinhibited and dephosphorylates WEE1, preventing its degradation and activating the G2/M checkpoint by WEE1. Proteins whose activity leads to cell cycle arrest are shown in red and proteins whose activity leads to cell cycle progression are shown in blue. Red DNA strands indicate newly synthesized DNA. Pointed arrows indicate action on that protein (phosphorylation) and blunted arrows indicate inhibition. BLM, Bloom syndrome protein; HRR, homologous recombination repair pathway; ICL, inter-strand crosslink pathway; MCM, minichromosome maintenance; NER, nucleotide excision repair pathway; RRM2, ribonucleoside-diphosphate reductase subunit M2; WRN, Werner syndrome ATP-dependent helicase.
