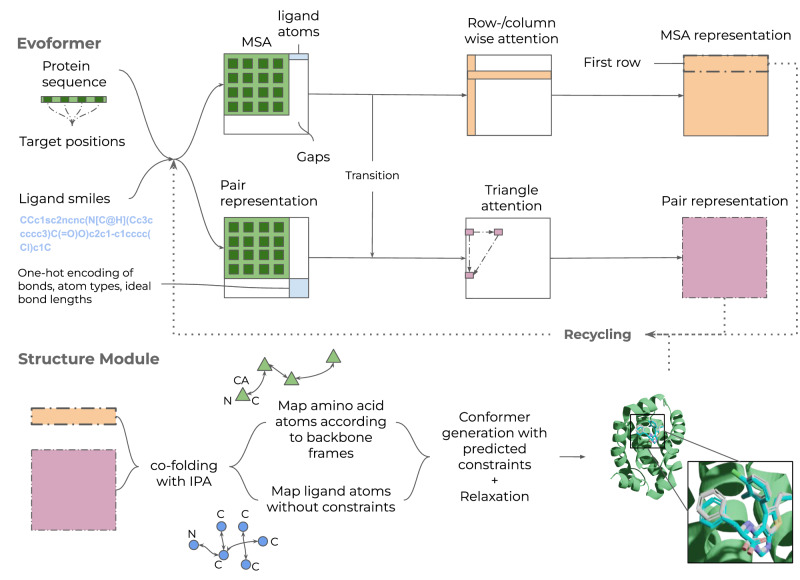
Fig. 1. Description of Umol.
An Evoformer network that processes both protein and ligand atom information. There are 48 Evoformer blocks. The protein is represented by a multiple sequence alignment (MSA) and optional target positions in the protein (pocket, Cβs within 10 Å from the ligand) are also defined. When target positions are not known, this information can simply be left out. A SMILES string represents the ligand. Two different tracks are present. The top track processes the MSA (gaps for the ligand) and the bottom track processes pairwise connections within and between the protein and ligand. The resulting representations are fed into the structure module (8 blocks) which produces a 3D structure of the protein-ligand complex. The entire network is trained end-to-end and the representations and predicted atom positions are recycled to refine the final structure. An example for PDB ID 7NB4 (ligand RMSD = 0.57) is shown with the predicted protein in green, the predicted ligand in blue and the native ligand in grey. The ligands are coloured by atomic types (blue=nitrogen, red=oxygen, remaining=carbon). This example was predicted using pocket information.