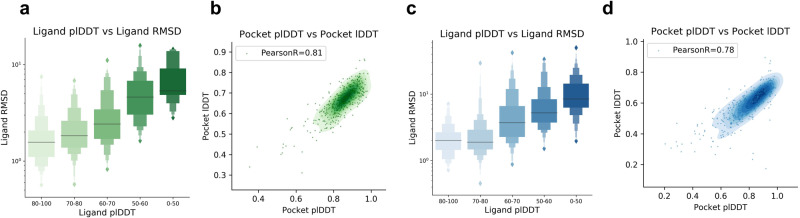
Fig. 3. Confidence metrics and accuracy.
a Ligand plDDT vs ligand RMSD for structures predicted with Umol-pocket (n = 428). The success rate (ligand RMSD ≤ 2 Å) for each bin is 80-100: 72.3%, 70-80: 58.2%, 60-70: 34.4%, 50-60: 5.3%, 0-50: 0.0%. The centre boxes encompass data quartiles and horizontal lines mark the medians for each distribution with min/max values marked by diamonds. b Protein pocket (all CBs within 10 Å from any ligand atom) predicted lDDT (plDDT) vs pocket lDDT for structures predicted with Umol-pocket (n = 428) as a density plot with each datapoint represented as a green scatter. The Pearson correlation coefficient is 0.81, suggesting a strong relationship between the two. c Ligand plDDT vs ligand RMSD for structures predicted with Umol (n = 428). The centre boxes encompass data quartiles and horizontal lines mark the medians for each distribution with min/max values marked by diamonds. The success rate (ligand RMSD ≤ 2 Å) for each bin is 80–100: 50.0%, 70–80: 52.6%, 60–70: 16.7%, 50–60: 1.8%, 0–50: 1.2%. At a plDDT threshold of >85, the SR is 80% for Umol without pocket information, enabling the selection of blind predictions with high confidence. d Protein pocket predicted lDDT (plDDT) vs pocket lDDT for structures predicted with Umol (n = 428) as a density plot with each datapoint represented as a green scatter. The Pearson correlation coefficient is 0.78.