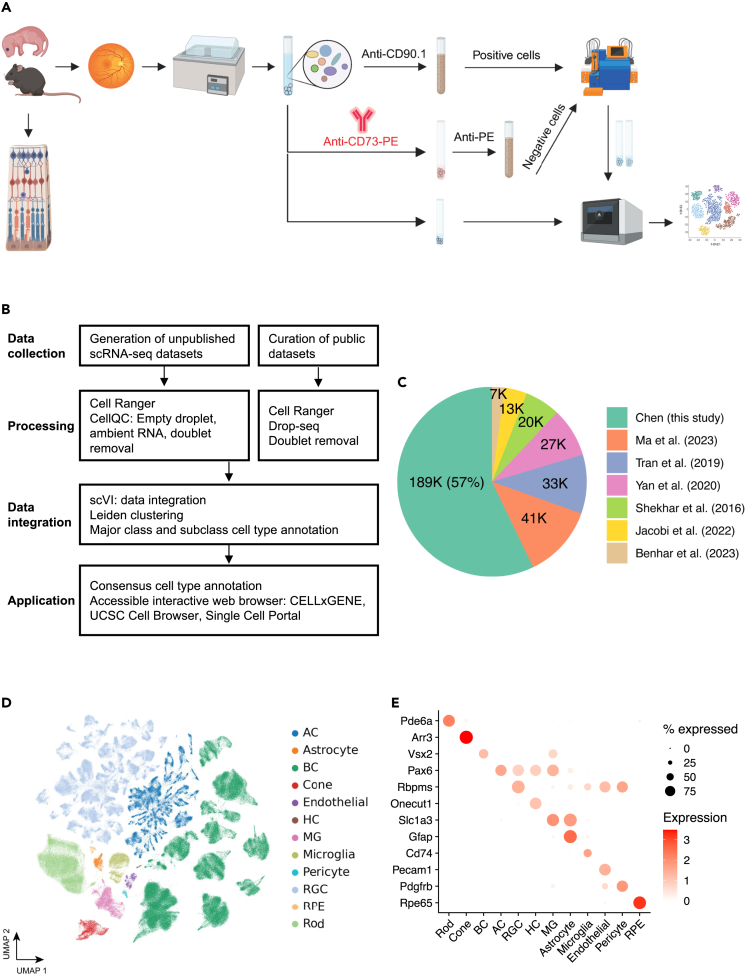
Figure 1.
Overview of single cell atlas of the mouse retina
(A) The workflow for generating unpublished scRNA-seq datasets. The data generation process involved using mice aged from P14 to 12 months. Following retina dissection and cell dissociation, single cells were enriched using autoMACS with Anti-CE73-PE antibodies or Anti-CD90.1 beads for specific amacrine, bipolar, and retinal ganglion cells. Subsequently, 10X single-cell RNA sequencing was performed on both the unenriched and enriched single cells. The retained single cells were then utilized in downstream atlas construction.
(B) The integrated analysis workflow for constructing the MRCA. To construct a comprehensive unified single-cell reference of the mouse retina, we generated 16 unpublished scRNA-seq samples of the mouse retina and incorporated four curated public datasets to enhance specific amacrine, bipolar and retinal ganglion cells. The collected data were processed using the Cell Ranger and CellQC pipeline to produce feature count matrices. Feature counts were then processed to remove estimated empty droplets, ambient RNA, and doublets. The retained cells were integrated using scVI to eliminate batch effects across samples. The trained low-dimensional embeddings were used to calculate cell dissimilarities and identify clustering through a two-level clustering approach. Major class and subclass cell types were annotated using canonical marker genes and public labeling. To facilitate user-friendly access and exploration, the MRCA was deployed on accessible interactive web browsers, including CELLxGENE, UCSC Cell Browser, and Single Cell Portal.
(C) Pie chart displaying the percentage of cells contributed by each dataset used in the MRCA.
(D) UMAP visualization of the MRCA colored by major classes.
(E) Dot plot illustrating the expression of canonical markers for major classes.