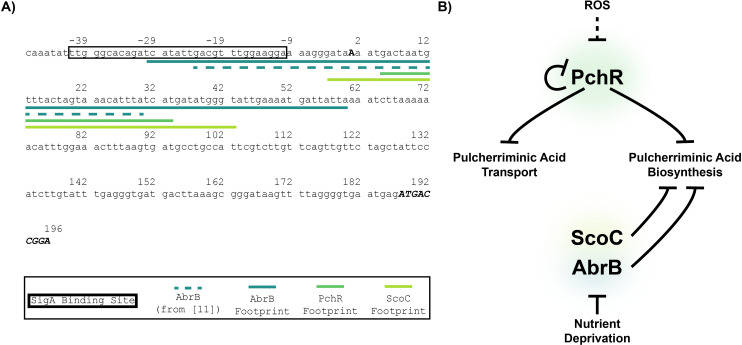
Fig 7. Model of Pulcherrimin Regulation by ScoC, AbrB, and PchR.
A) Promoter sequence of yvmC. The putative SigA binding site is marked with a box and the transcriptional start site is bold and upper case. Regions protected by transcription factors in the DNase I footprinting assays are provided by colored lines under the DNA sequence. The dashed line represents the sequence in the region pulled down by AbrB in vivo by Chumsakul et al [11]. The translation start site and the first 9 base pairs of yvmC are bold, italicized, and in upper case. B) The MarR-family transcription factor PchR negatively regulates its own expression as well as the genes needed for pulcherriminic acid biosynthesis (yvmC and cypX) and transport (yvmA). Our work identifies members of the transition state regulators, ScoC and AbrB, as direct negative regulators of pulcherrimin biosynthesis by inhibiting expression of the biosynthesis gene yvmC. The location of the protection areas in the yvmC promoter suggest all transcription factors act to inhibit binding and/or progression of RNAP. Nutrient deprivation relieves the repressive effect of the transition state regulators ScoC and AbrB. ROS relieves PchR-mediated repression of the pulcherrimin biosynthesis and transporter operons in B. licheniformis, however this has yet to be experimentally verified in B. subtilis [46].