Figure 6.
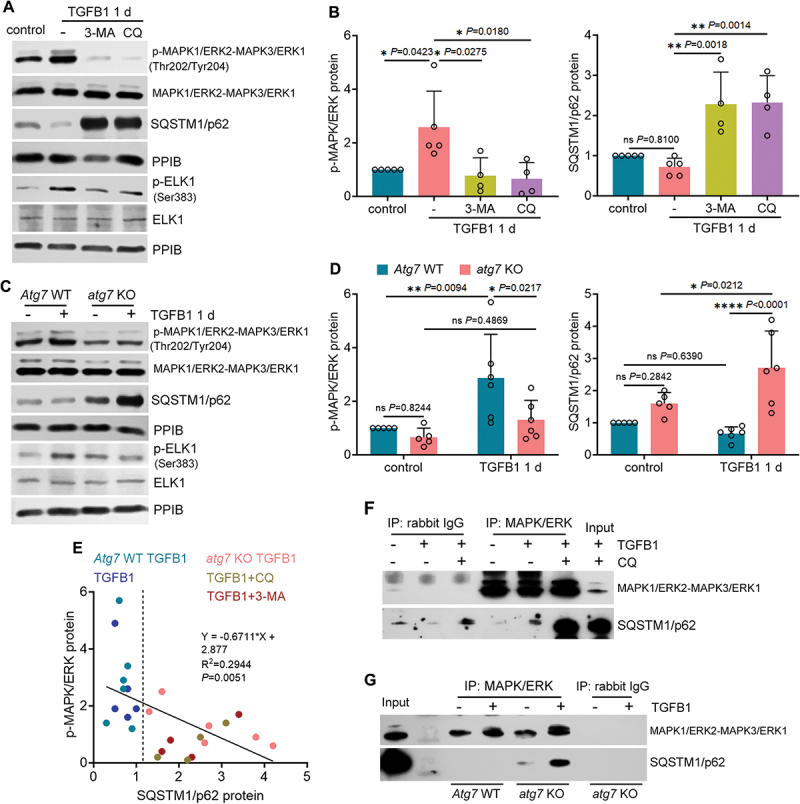
SQSTM1/p62 interacts with MAPK1/ERK2-MAPK3/ERK1 to block its activation by TGFB1 in autophagy-deficient mouse proximal tubular cells. (A and B) subconfluent BUMPT cells were exposed to 5 ng/ml TGFB1 in serum-free DMEM for 1 day alone or with 20 µM CQ or 5 mM 3-MA. Control cells were kept in serum-free medium without TGFB1. Cells were collected for immunoblot of p-MAPK1/ERK2-MAPK3/ERK1 (Thr202/Tyr204), MAPK1/ERK2-MAPK3/ERK1, SQSTM1/p62, p-ELK1 (Ser383) and ELK1 (n = 5 experiments). (C and D) subconfluent WT and atg7 KO cells were exposed to 5 ng/ml TGFB1 in serum-free DMEM for 1 day. Control cells were kept in serum-free medium without TGFB1. Cells were collected for immunoblot of p-MAPK1/ERK2-MAPK3/ERK1 (Thr202/Tyr204), MAPK1/ERK2-MAPK3/ERK1, SQSTM1/p62, p-ELK1 (Ser383) and ELK1 (n = 5 experiments). (E) correlation analysis of protein fold changes between p-MAPK1/ERK2-MAPK3/ERK1 (Thr202/Tyr204) and SQSTM1/p62. (F) co-IP of MAPK1/ERK2-MAPK3/ERK1 and SQSTM1/p62 in BUMPT cells (n = 3 experiments). (G) co-IP of MAPK1/ERK2-MAPK3/ERK1 and SQSTM1/p62 in WT and atg7 KO cells (n = 3 experiments). Data in (B) and (D) are presented as mean ± SEM. For statistics, one-way ANOVA with multiple comparisons was used for (B). Two-way ANOVA with multiple comparisons was used for (D). Pearson correlation analysis followed by simple linear regression was used for (E).
