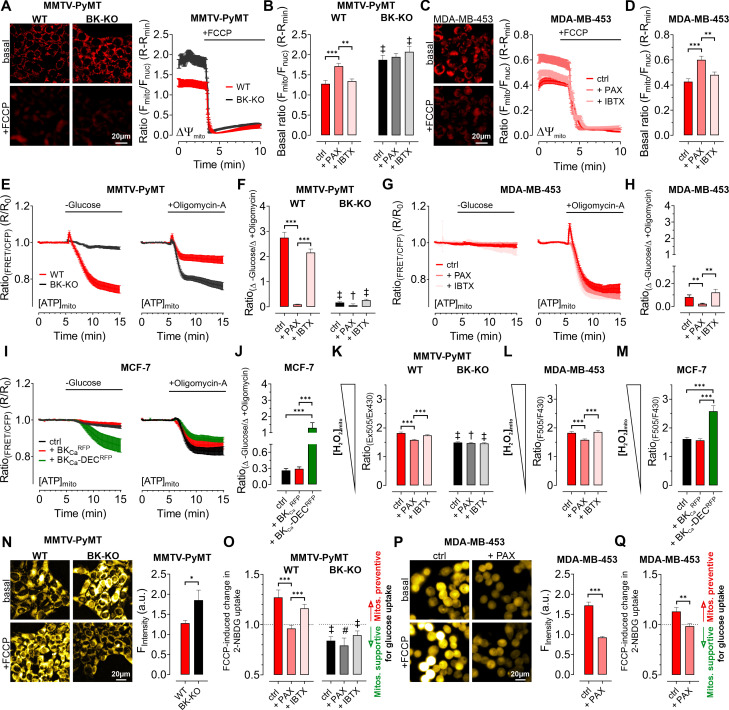
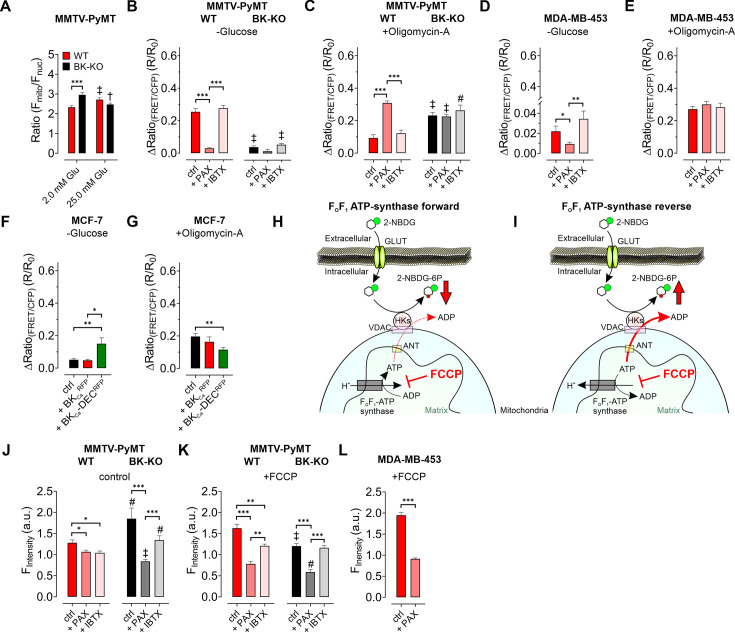
Figure 4. Expression of BKCa modulates mitochondrial function and glucose uptake of BCCs.
(A – D) Representative fluorescence images and -ratios (Fmito/Fnuc) over-time (A, C), and corresponding statistics ± SEM (B, D) representing ΔΨmito of TMRM-loaded MMTV-PyMT WT and BK-KO (A, B) and MDA-MB-453 cells (C, D) under basal conditions (A, C, upper images) and upon administration of FCCP for mitochondrial depolarization (A, C, lower images). (E – J) [ATP]mito dynamics ± SEM over-time of MMTV-PyMT WT and BK-KO cells (E), MDA-MB-453 cells (G) and MCF-7 cells (I) in response to extracellular glucose removal (left panels) or upon administration of Oligomycin-A (right panels). (F, H) and (J) show changes of [ATP]mito induced by glucose removal to Oligomycin-A administration ± SEM, under control conditions, or in the presence of paxilline or iberiotoxin (F, H), or upon expression of BKCaRFP or BKCa-DECRFP (J). (K – M) Basal mitochondrial H2O2 concentrations ± SEM of MMTV-PyMT WT (K, left), BK-KO (K, right), MDA-MB-453 (L) and MCF-7 cells (M), either under control conditions, in the presence of paxilline or iberiotoxin (K, L), or upon expression of BKCaRFP or BKCa-DECRFP (M). (N, P) Representative fluorescence wide-field images (left) and corresponding statistics ± SEM (right) of MMTV-PyMT WT (N, left images and red bars) and BK-KO cells (N, right images and black bars) or MDA-MB-453 cells (P) incubated with 2-NBDG, either in the absence (upper images) or presence of FCCP (lower images). (O, Q) Average ± SEM of FCCP induced change in 2-NBDG uptake of MMTV-PyMT WT (O, left) and BK-KO cells (O, right), or MDA-MB-453 cells (Q) either under control conditions, or in the presence of paxilline or iberiotoxin. Values above 1 indicate that mitochondria prevent, values below 1 that mitochondria support glucose uptake. N (independent experiments) / n (cells analyzed) = (A, B): 4/75 WT ctrl, 4/90 WT +PAX, 4/86 WT +IBTX, 4/91 BK-KO ctrl, 4/89 BK-KO +PAX, 4/100 BK-KO +IBTX, (C, D): 4/113 ctrl, 4/97+PAX, 4/103+IBTX, (E, F): [-Glucose]: 8/55 WT ctrl, 6/45 WT +PAX, 7/27 WT +IBTX, 8/65 BK-KO ctrl, 6/57 BK-KO +PAX, 7/28 BK-KO +IBTX, [+Oligomycin-A]: 11/52 WT ctrl, 7/53 WT +PAX, 7/34 WT +IBTX, 8/87 BK-KO ctrl, 6/35 BK-KO +PAX, 5/45 BK-KO +IBTX. (G, H): [-Glucose]: 5/14 ctrl, 3/13+PAX, 5/13+IBTX, [+Oligomycin-A]: 5/33 ctrl, 3/21+PAX, 8/27+IBTX, (I, J): [-Glucose]: 6/48 ctrl, 5/23+BKCaRFP, 5/20+BKCa-DECRFP, [+Oligomycin-A]: 5/27 ctrl, 5/23+BKCaRFP, 5/37+BKCa-DECRFP, (K): 3/33 WT ctrl, 4/51 WT +PAX, 4/54 WT +IBTX, 4/55 BK-KO ctrl, 4/51 BK-KO +PAX, 4/54 BK-KO +IBTX, (L): 4/31 ctrl, 4/39+PAX, 4/31+IBTX, (M): 4/29 ctrl, 4/17+BKCaRFP, 4/21+BKCa-DECRFP, (N – Q): 4 for all. *p≤0.05, **p≤0.01, ***p≤0.001, Kruskal-Wallis test followed by Dunn’s MC test (B, D, F, H, J, K, O), Brown-Forsythe and Welch ANOVA test followed by Games-Howell’s MC test (L, M), Mann-Whitney test (N), Unpaired t-test (P) or Welch’s t-test (Q). #p≤0.05, †p≤0.01, ‡p≤0.001, to respective WT condition, Mann-Whitney test (B, F) +PAX and+IBTX in (K) ctrl in (O), Unpaired t-test (ctrl in K) +PAX and+IBTX in (O).