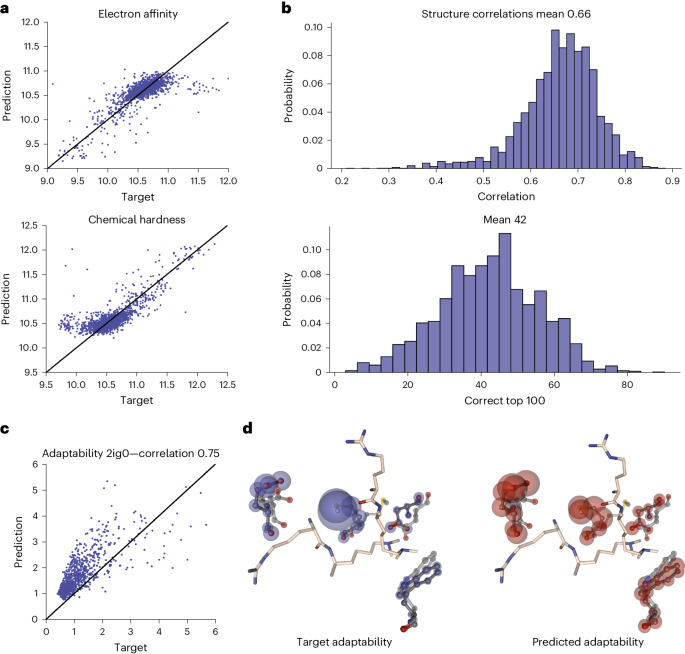
Fig. 5. Performance of the AI baseline models.
a, Scatter plot of the predicted against target values of chemical hardness and electron affinity. The AI baseline models to predict QM properties have a high correlation of 0.75 and 0.77 for electron affinity and chemical hardness, respectively. b, Adaptability is a measure of the per-atom conformational plasticity of the protein. A histogram of the correlation and the correct top 100 predictions of the adaptability for all structures in the test set are given. An overall mean correlation of 0.66 can be achieved and the mean top 100 accuracy was 0.42 for the adaptability predictions (MD). c, Scatter plot for the adaptability result (as in a) of example structure 2IG0. The predicted values are more narrowly distributed than the actual values, but the general trend is correct, as shown by a high correlation value of 0.75. d, The adaptability of the residues in the protein pocket highly deviates between the amino acids. The AI model predicts the adaptability given in blue-shaded (target) and red-shaded (AI-predicted) spheres. The radius is scaled according to the adaptability value. The model can correctly identify the rigid residues (small spheres) but also the amino acids with high flexibility. Color and character keys: N, blue, nitrogen; S, yellow, sulfur; O, red, oxygen; C, beige for ligand atoms and black for protein atoms, carbon.