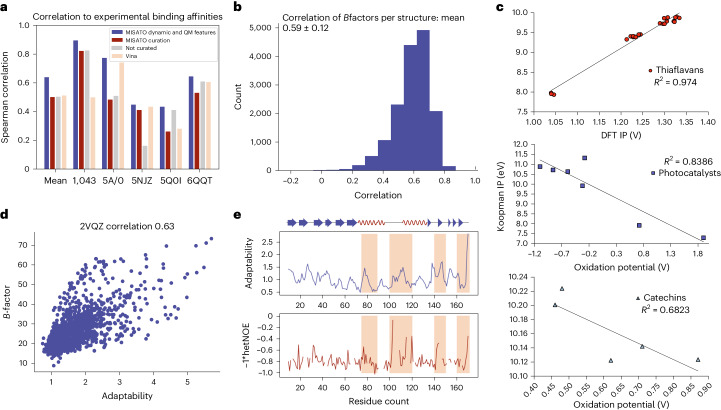
Fig. 6. Experimental validation of QM calculations, MD traces and AI models.
a, Spearman correlation of the affinity GNN model on the binding affinity benchmark including MISATO features and without features. Moreover, the results using Vina and non-curated complexes (original PDBbind) are shown. We achieved a consistently better performance including QM charges and MD adaptabilities as MISATO features across the affinity benchmark when compared with all other approaches. b, Histogram of the correlation of experimental B factors from X-ray crystallography experiments with RMSF calculations from the MD simulations in MISATO. A correlation of 0.59 over all structures was achieved. c, High correlations of calculated Koopmans ionization potentials (IP) from ULYSSES with DFT ionization potentials (upper panel) and experimental oxidation potentials (middle and lower panels) were found for different molecule families. d, The cap-binding domain of influenza virus polymerase as a model system for experimental validation of the predicted adaptability. Values given by our AI model had a high correlation of 0.63 against the experimentally determined B factors (which, despite characterizing atom thermal vibration, usually indicates flexibility). e, Results of the hetNOE experiments of the cap-binding domain of influenza virus polymerase indicating flexibility of the protein chain were in high accordance with the results of the adaptability model (indicated using shaded regions).