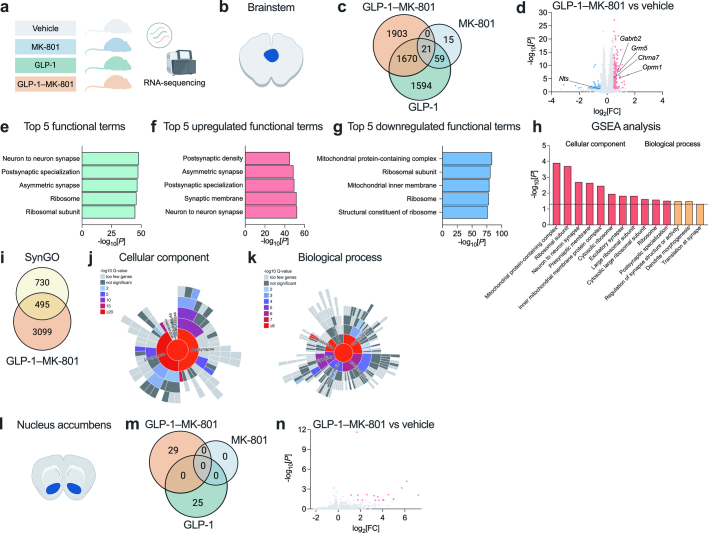
Extended Data Fig. 12. Bulk RNA sequencing of brainstem and nucleus accumbens after treatment with GLP-1–MK-801.
a-n, Brain nuclei for bulk RNA sequencing of the brainstem came from mice in Fig. 3h, i and Extended Data Fig. 8 treated with 100 nmol kg−1 MK-801 (n = 8 mice), 100 nmol kg−1 GLP-1 (n = 8 mice), 100 nmol kg−1 GLP-1–MK-801 (n = 8 mice) or vehicle (isotonic saline, n = 8 mice) for 5 days. a, Schematic. b, Schematic highlighting the brainstem. c, Venn diagram of differentially expressed genes in response to treatments. d, Volcano plot for differentially expressed genes in response to GLP-1–MK-801. e, Top five most enriched functional terms for the 3594 differentially expressed genes between GLP-1–MK-801 and vehicle. f, Top five most enriched functional terms for the 2076 differentially expressed genes that were upregulated in response to GLP-1–MK-801 treatment relative to vehicle. g, Top five most enriched functional terms for the 1518 differentially expressed genes that were downregulated following GLP-1–MK-801 treatment relative to vehicle. h, Top eleven cellular component and top three biological process functional terms identified using gene set enrichment (GSEA) analysis. i, Venn diagram showing the overlap with the SynGO database and the 3594 differentially expressed genes in response to GLP-1–MK-801. j, Sunburst plot of differentially expressed genes belonging to cellular component functional terms computed with SynGO. Enrichment is colour coded by Q value for the functional terms. All level terms identified have been labelled. k, Sunburst plot of differentially expressed genes belonging to biological process functional terms computed with SynGO. Enrichment is colour coded by Q value for the functional terms. All level terms identified have been labelled. l, Schematic highlighting the nucleus accumbens. m, Venn diagram of differentially expressed genes in response to treatments. n, Volcano plot for differentially expressed genes of GLP-1–MK-801. Benjamini-Hochberg adjusted P (c-n). Detailed statistics are in Supplementary Table 1. The diagrams in a-c, i, l and m were created using BioRender.