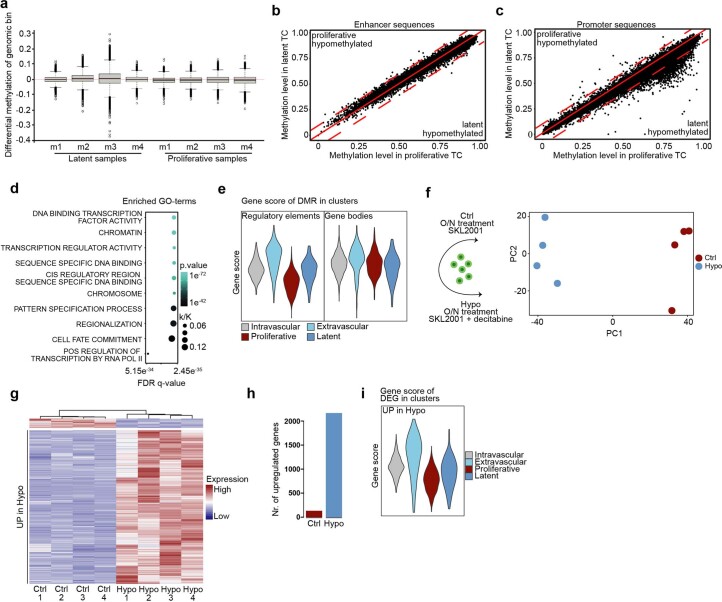
Extended Data Fig. 10. Latent tumor cell hypomethylation in promoter sequences and gene bodies drives plasticity.
a, Boxplot reflecting the genome wide differential methylation level. Genome was binned in parts of 2000 bp. Methylation fractions were calculated for each bin by the ratio of methylated to total CpG island per bin. Methylation levels were assessed by subtracting the median methylation fraction of a bin across all samples from each individual bin. Red line indicates median methylation fraction of genomic bin. Black lines indicate median for each sample. Top and bottom of box show 25th and 75th percentile, respectively. Whiskers represent the 1.5 interquartile range. b, Scatter plot of methylation level (fraction of methylated CpG islands) for enhancer or c, promoters in latent TC and proliferative TC. Red line indicates no differences in methylation, dotted red lines indicate thresholds for >10% differences in methylation. d, Enriched GO-terms for hypomethylated genes in latent TC. k/K, overlap of queried gene list and GO-term associated genes. FDR, false discovery rate. p values were computed by permutation. e, Scored gene expression of genes linked to hypomethylated regulatory elements in latent tumor cells (as identified in panels b, c) (left panel) or gene bodies (Fig. 5g) (right panel) in the scRNAseq TC dataset split by cell clusters. f, Schematic of RNAseq experiment. Tumor cells were either treated with SKL2001 (Ctrl) or SKL2001 and decitabine (Hypo) overnight. PCA plot of sequencing results (right panel). n = 4 replicates. g, Row normalized and row clustered heatmap of differentially expressed genes between Ctrl and Hypo samples. Genes were considered significantly regulated for log 2 fold changes > 0.58 and adjusted p value < 0.05. p values were computed in DESeq2 using Wald test. h, Bar plot displaying number of upregulated genes for each treatment group (full set of DEG is displayed in Supplementary Table 9). i, Scored gene expression of Hypo gene signature from the RNAseq experiment in the scRNAseq dataset split by cell clusters.