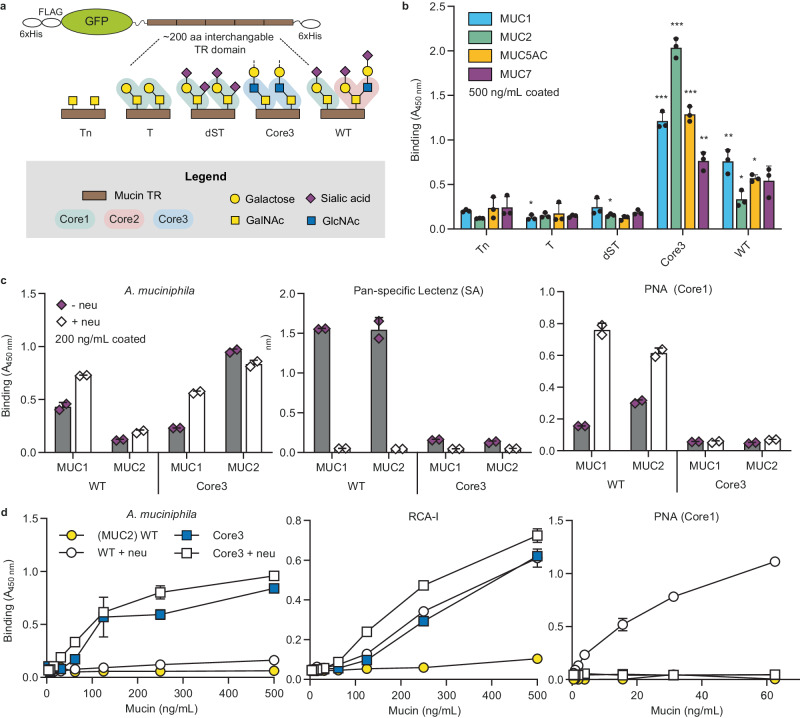
Fig. 2. ELISA binding assays of pasteurized A. muciniphila with purified glycoengineered human mucin TR reporters.
a Graphic depiction of the mucin reporters and the O-glycoforms analyzed. The mucin TR reporters were produced in cells engineered as follows: Tn (KO C1GALT1), T (KO GCNT1, KO ST6GALNAC2/3/4, KO ST3GAL1/2), dST (KO GCNT1, KI ST6GALNAC2/3/4), core3 (KO COSMC, KI B3GNT6). The WT HEK293 cells produce a mixture of core1 (ST) and core2 O-glycans as illustrated. b Binding of pasteurized A. muciniphila (5 × 108 CFU/mL) to isolated mucin TR reporters with five different glycoforms as indicated. Binding to Tn of respective TR domain serves as a reference for statistical analysis. Bars represent the mean ± SD of three biological replicates. A Student’s two-sided t-test was performed to assess differences in means between conditions. *p < 0.05, **p < 0.01, ***p < 0.001. c Binding to mucin reporters (200 ng/mL) with or without pretreatment with Clostridium perfringens neuraminidase (20 mU overnight) (A. muciniphila 1 × 109 CFU/mL) with control binding of pan-specific Lectenz (2 µg/mL) and PNA (0.1 µg/mL). One representative experiment is shown, with bars representing the mean ± SD of 2 technical replicates. d Binding of pasteurized A. muciniphila (5 × 108 CFU/mL) to varying concentrations of WT and Core3 MUC2 reporters pretreated with and without C. perfringens neuraminidase. Binding of RCA-I (0.05 μg/mL) and PNA (0.1 µg/mL) as controls. One representative experiment is shown, representing the mean ± SD of 2 technical replicates. Source data are provided as a Source Data file.