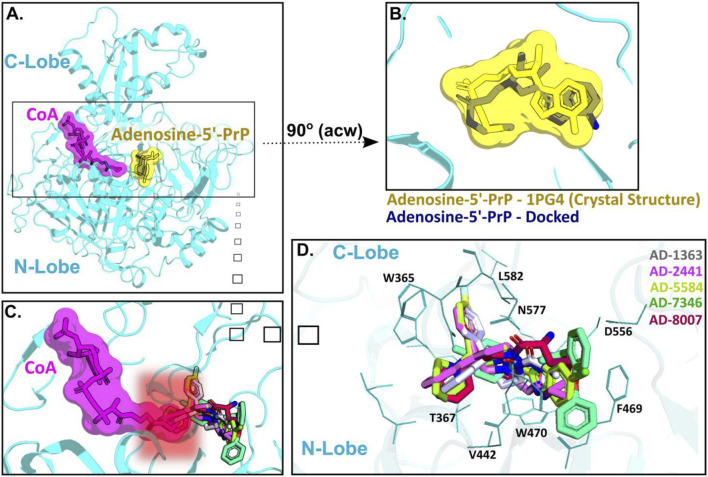
FIGURE 4.
AD-2441 and its analogs are predicted to bind ACSS2 within the nucleotide-binding pocket and stabilized by various hydrophobic and polar contacts and aromatic stacking interactions. Docking calculations were performed into a homology model (using swissmodel.expasy.org) (Waterhouse et al., 2018; Studer et al., 2020) of ACSS2 based on the crystal structure of Salmonella enterica acetyl-CoA synthetase in complex with cAMP and Coenzyme A (PDB: 5JRH). (A) Homology model of ACSS2 with superimposed CoA (purple) and Adenosine-5′-propylphosphate (yellow, extracted from PDB: 1PG4). (B) Superimposed crystal structure of Adenosine-5′-propylphosphate (yellow, extracted from PDB: 1PG4) and docked (DiffDock and Flare version 5 minimized) Adenosine-5′-propylphosphate pose (blue). (C) All docked compounds are predicted to bind within the Adenosine-5′-propylphosphate site and additionally sterically interfere with CoA (purple) binding indicated by the red helo. (D) Close-up view of binding poses of AD-1363, AD-2441, AD-5584, AD-7346, and AD-8007 between the C- and N-Lobe of ACSS2.