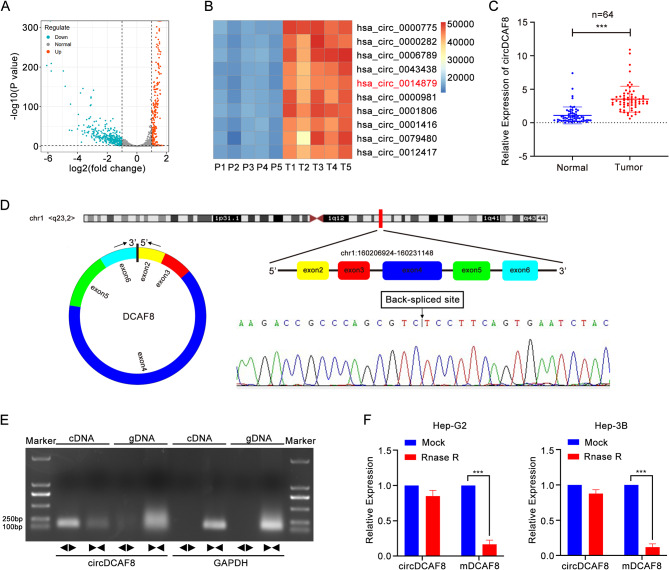
Fig. 1.
Selection and identification of circDCAF8. A The volcano plot of DECs in GSE94508. B A heatmap of the top 10 upregulated circRNAs in 5 paired samples of HCC. C Relative expression of circDCAF8 in human HCC tissues and paired adjacent nontumor tissues of 64 patients was determined by qRT-PCR. D Sanger sequencing of the annotated genomic region of circDCAF8 was performed to confirm the Back-spliced site of circDCAF8. E The divergent primers detected circDCAF8 in cDNA but not in gDNA by agarose gel electrophoresis. GAPDH was used as a negative control. F qRT–PCR analysis for the expression of circDCAF8 and mDCAF8 after treatment with RNase R in Hep-G2 and Hep-3B cells. Data are representative of three independent experiments and are presented as means ± SDs. (*p < 0.05; **p < 0.01; ***p < 0.001)