SUMMARY
Histone lysine acylation including acetylation and crotonylation plays a pivotal role in gene transcription in health and diseases. However, our understanding of histone lysine acylation has been limited to gene transcriptional activation. Here we report that histone H3 lysine 27 crotonylation (H3K27cr) directs gene transcriptional repression rather than activation. Specifically, H3K27cr in chromatin is selectively recognized by the YEATS domain of GAS41 in complex with SIN3A-HDAC1 co-repressors. Proto-oncogenic transcription factor MYC recruits GAS41/SIN3A-HDAC1 complex to repress genes in chromatin including cell cycle inhibitor p21. GAS41 knockout or H3K27cr binding depletion results in p21 de-repression, cell cycle arrest and tumor growth inhibition in mice, explaining a causal relationship between GAS41 and MYC gene amplification and p21 down-regulation in colorectal cancer. We study suggests that H3K27 crotonylation signifies a previously unrecognized, distinct chromatin state for gene transcriptional repression in contrast to H3K27 trimethylation for transcriptional silencing and H3K27 acetylation for transcriptional activation.
Graphical Abstract
INTRODUCTION
Lysine crotonylation is closely related to acetylation in chemical structure and takes place at major acetylation sites in histones.1 Yet, histone lysine crotonylation is recognized by structurally distinctive YEATS domain rather than the well-known acetyl-lysine binding bromodomain,2,3 suggesting different underlying histone biology concerning these distinct lysine acylation states. Human genome encodes four YEATS domain proteins AF9, ENL, YEATS2, and GAS41 (a.k.a. YEATS4), found in chromatin remodeling and histone acetyltransferase complexes in regulation of chromatin structure, histone lysine acylation, gene transcription, and DNA damage response.4 AF9 participates in DOT1L-mediated H3K79 methylation in gene transcription.5,6 AF9 or ENL act as oncogenic drivers in acute myeloid leukemia via YEATS domain/acetyl-lysine binding for active gene transcription.7,8 Mutations in the ENL YEATS domain favor clinical development of Wilms tumor.9,10 Disruption of YEATS2 YEATS domain/histone binding reduces histone H3 lysine 9 acetylation at gene promoter by ATAC complex and down-regulates expression of ribosomal protein genes critical for non-small cell lung cancer.11 Additionally, a yeast YEATS domain protein Taf14 was reported to act in histone crotonylation-mediated metabolic flux through transcriptional repression of energy-demanding gene expression.12
Distinct from AF9 and ENL, GAS41 that consists of the YEATS domain at the N-terminus and the coiled-coil motif at the C-terminus (Figure 1A) has been reported for gene transcriptional repression with a poorly understood mechanism.13 Originally identified in glioblastoma multiforme cells in early glioma tumor development,14 GAS41 is known for its gene amplification in colorectal cancer,15 gastric cancer,16 non-small cell lung cancer,17 glioblastomas,18 and pancreatic cancer.19 GAS41 was reportedly associated via unknown mechanism with TIP6020 and SRCAP21 modeling complexes, as well as transcription factors MYC, INI1,22 TFIIF23 and AP-2β,24 leukemia fusion protein MLL-AF10,25 and cancer-related TACC1,26 and implicated in tumorigenesis27 through inhibiting p53-p21 tumor suppression activity28 or activating Wnt/β-catenin signaling pathway.16 Recent studies show that GAS41 co-localizes with lysine-acetylated histones for gene transcription, and depletion of GAS41 association with histone variant H2A.Z results in suppression of cancer cell growth and survival in non-small cell lung cancer cells.29,30 In this study, we report that histone H3 lysine 27 crotonylation (H3K27cr) acts to direct gene transcriptional repression rather than activation through a previously unrecognized mechanism that entails selective interaction with the YEATS domain of GAS41, which is recruited by MYC to target genes in chromatin and works with the SIN3A-HDAC1 co-repressors for gene transcriptional repression.
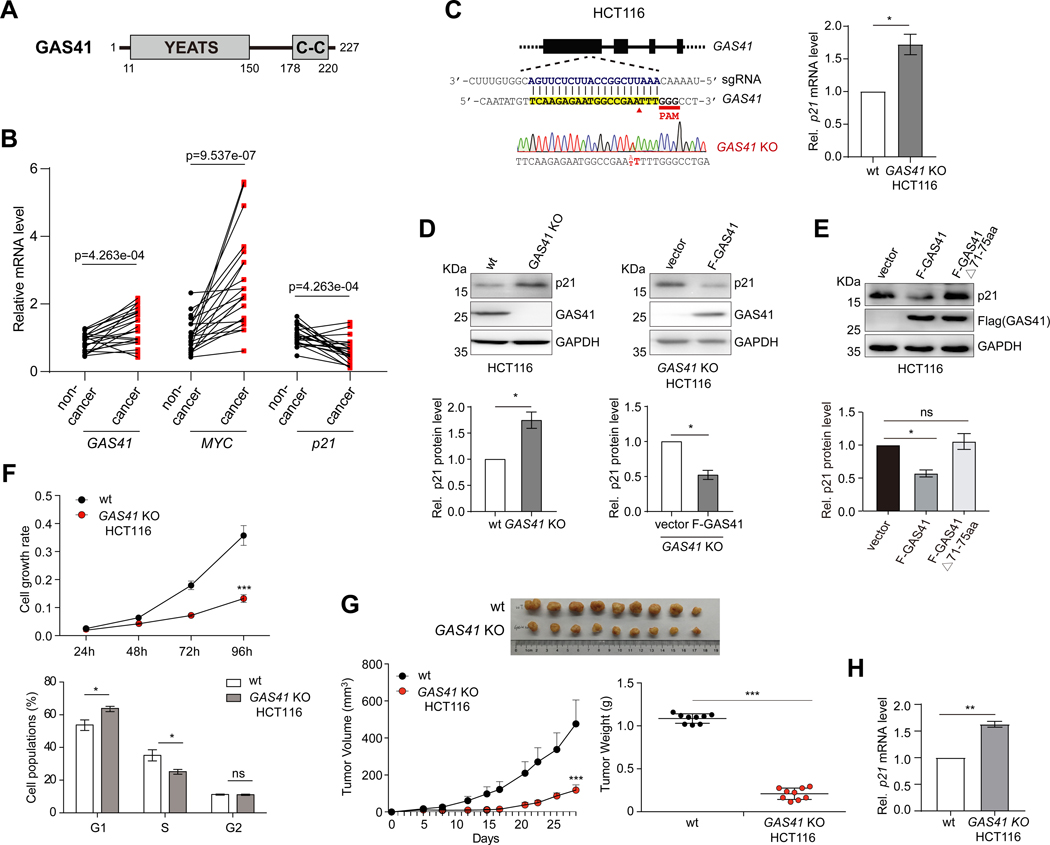
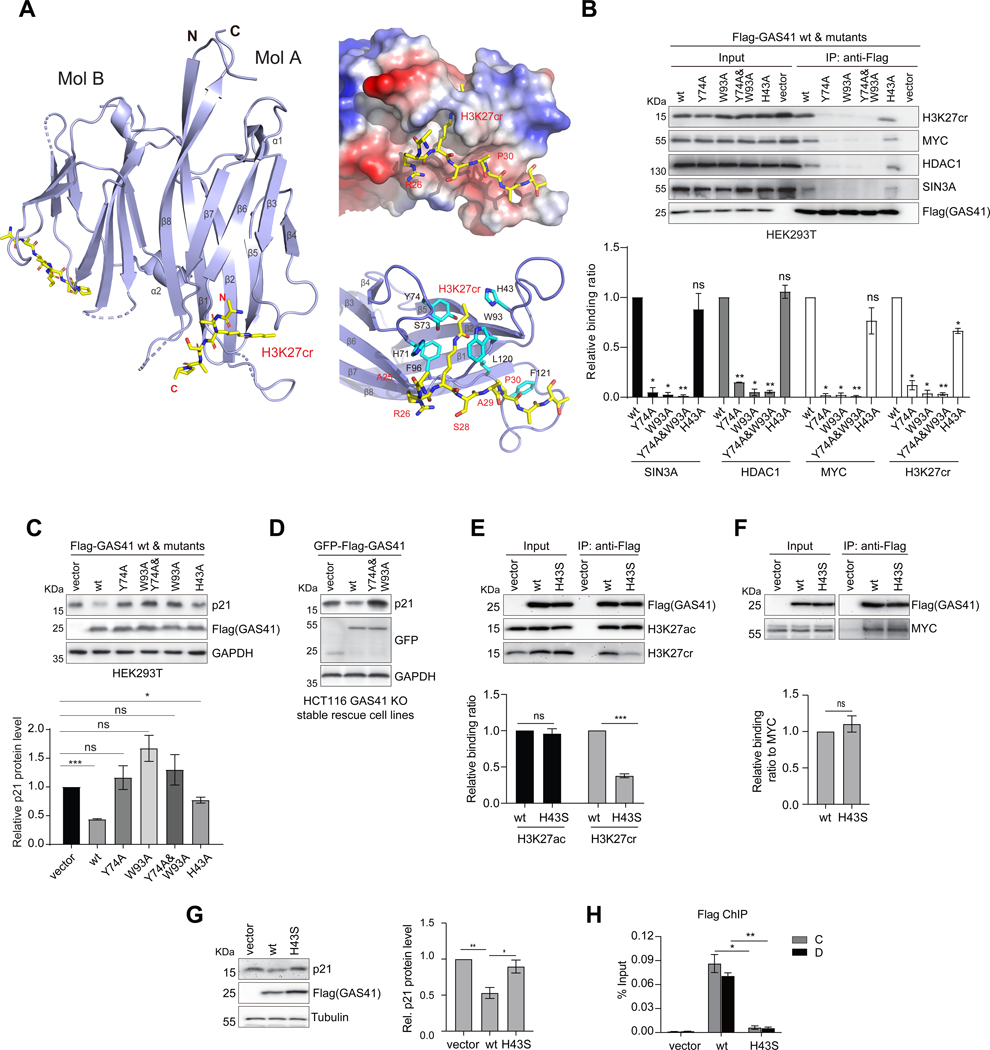
Figure 1. GAS41 contributes to p21 repression in tumor cell proliferation.
(A) Scheme depicting protein functional domain organization of GAS41, consisting of the YEATS domain at the N-terminus and the coiled-coil motif at the C-terminus.
(B) Before-after plot showing mRNA expression levels of GAS41, p21 and MYC in 21 pairs of cancer vs. adjacent non-cancerous tissues of human colorectal cancer patients. GAS41, MYC or p21 expression level in non-cancer tissue of #1 was set as 1. Statistical significance (p value, as indicated) was calculated with Wilcoxon signed-rank test (paired samples Wilcoxon test).
(C) Scheme depicting guide RNA design targeting GAS41 start codon for generating HCT116 GAS41 knockout (KO) cells using CRISPR/Cas9 method. Right, RT-qPCR analysis of p21 expression in wild type (wt) and HCT116 GAS41 KO cells. n=3.
(D) Western blot analysis and quantification of p21 and GAS41 expression in HCT116 GAS41 KO cells with or without transfection of Flag-GAS41 using antibodies as indicated. n=3.
(E) Western blot analysis and quantification of p21 expression in HCT116 cells transiently transfected with a Flag vector or a vector expressing Flag-GAS41 wt or YEATS domain mutant (Δ71–75). n=3.
(F) Cell growth rate of HCT116 wt and GAS41 KO cells as assessed by MTT assay. Shown were mean ± s.e.m. from 6 technical repeats. Lower, cell cycle analysis of wt and HCT116 GAS41 KO cells by fluorescence-activated cell sorter analysis. Relative distributions of cells in different cell cycle phases, as analyzed using FlowJo software. n=3.
(G) Effect of GAS41 knockout on tumor volume and weight in mice. 1×106 HCT116 wt or GAS41 KO cells were injected into the mammary fat pad of NOD-SCID mice. The tumor volume and weight were recorded for 29 days. Left, mean + s.d, Right, mean ± s.d. n=9.
(H) RT-qPCR analysis of p21 expression in the tumors derived from HCT116 wt and GAS41 KO cells in the mouse xenograft study. n=3. KO stands for knockout, wt, wild type, and F-GAS41, Flag-GAS41. All error bars throughout this study represent mean ± s.e.m. from indicated (n) biological repeats and statistical significance was calculated using two-tailed paired Student’s t-test unless noted otherwise. *P < 0.05, **P < 0.01, ***P < 0.001, ns, not significant.
See also Figure S1.
RESULTS
GAS41 Represses p21 Transcriptional Expression
To better understand GAS41 function in tumorigenesis, we analyzed and found that mRNA transcript levels of GAS41 and MYC in human colorectal cancer tissues are significantly higher than 21 paired adjacent non-cancerous tissues, while cell cycle inhibitor p21 showed consistently reduced mRNA expression in the cancer tissues than non-cancer tissues (Figures 1B, S1A and S1B). Close analysis revealed that 16 out of the 21 paired samples (76.2%) showed higher expression of GAS41, and 11 of the 16 samples (68.8%) showed decreased p21 (Figure S1A). The inverse correlation of GAS41 and p21 in transcriptional expression agrees with a previous report that GAS41 down-regulates p21 expression.13
To determine the role of GAS41 in p21 regulation, we generated GAS41 knockout (KO) cell lines from HCT116 and HEK293T cells using CRISPR/Cas9 with a designed guide RNA that targets the sequence following the GAS41 start codon (Figures 1C and S1C). Clones obtained from gene editing with non-homologous end joining carried two- or one-base insertion in HCT116 and HEK293T cells, respectively, resulting in premature stop codon and disruption of GAS41 translation. The GAS41 KO cells showed increased p21 expression, which was reinstated with ectopic expression of Flag-GAS41 (Figures 1D and S1D). In contrast, a YEATS domain deletion mutant (Flag-GAS41Δ71–75), designed to disrupt YEATS/acyl-lysine binding,31 was deficient in p21 repression in HCT116 cells (Figure 1E), implicating the importance of YEATS domain for GAS41 function in p21 regulation.
The GAS41 depletion caused cell growth reduction and cell cycle arrest at G1/S phase in HCT116 or HEK293T cells (Figures 1F and S1E), confirming GAS41’s role for cell proliferation. NOD/SCID mice injected with GAS41 KO HCT116 colorectal cancer cells produced much smaller tumors in size and weight than the control group mice injected with wild-type (wt) HCT116 cells (Figure 1G). The former group mice showed higher p21 expression in tumors than the latter (Figure 1H), corroborating GAS41 as an oncogene, responsible for repressing p21 expression.
GAS41 Binds Crotonylated-H3K27 for p21 Transcriptional Repression
Distinct from AF9 and ENL YEATS domains,6,7,32 we found that Flag-GAS41, ectopically expressed in HCT116 cells, prefers binding to biotinylated peptide of H3K27cr immobilized on streptavidin-magnetic beads over other peptides containing acetylated-H3K27 (H3K27ac), H3K9cr/ac, H3K14cr/ac, or H3K18cr/ac, as shown by Western blot analysis (Figure 2A), and this binding is abolished by point mutation of two key residues Y74 and W93 at the acyl-lysine binding pocket to Ala (Figure S2A; also see below). GAS41 YEATS domain binding preference for H3K27cr was confirmed by isothermal titration calorimetry (Figure S2B) and NMR 2D 1H-15N HSQC (heteronuclear single quantum coherence) spectroscopy (Figure S2C), showing that GAS41 favors H3K27cr over H3K27ac by about 2-fold in affinity (Kd of 22.9 μM vs. 51.5 μM). To further assess the binding preference for H3K27cr on the nucleosome, we performed microscale thermophoresis (MST) assay using nucleosome core particles (NCPs) with or without H3K27cr or H3K27ac modification. The affinity of GAS41 YEATS domain binding to NCPs is much higher than to the histone peptides with corresponding modifications (Figure S2D vs. Figure S2B), which agrees with the recent study of AF9 YEATS domain binding to NCPs vs. histone peptides.33 Importantly, the binding preference of the GAS41 YEATS domain to H3K27cr over H3K27ac is well retained with Kd = 158 nM for H3K27cr-NCP vs. Kd = 229 nM for H3K27ac-NCP and Kd = 913 nM for the unmodified NCP (Figure S2D). Our nucleosome binding study further illustrated that a H3K27cr peptide effectively blocked Flag-GAS41 binding to nucleosomal histone H3K27cr or H3K27ac in HEK293T cell lysate in a dose-dependent manner, whereas a H3K27ac peptide showed the inhibition only at higher concentrations (Figures 2B and S2E).
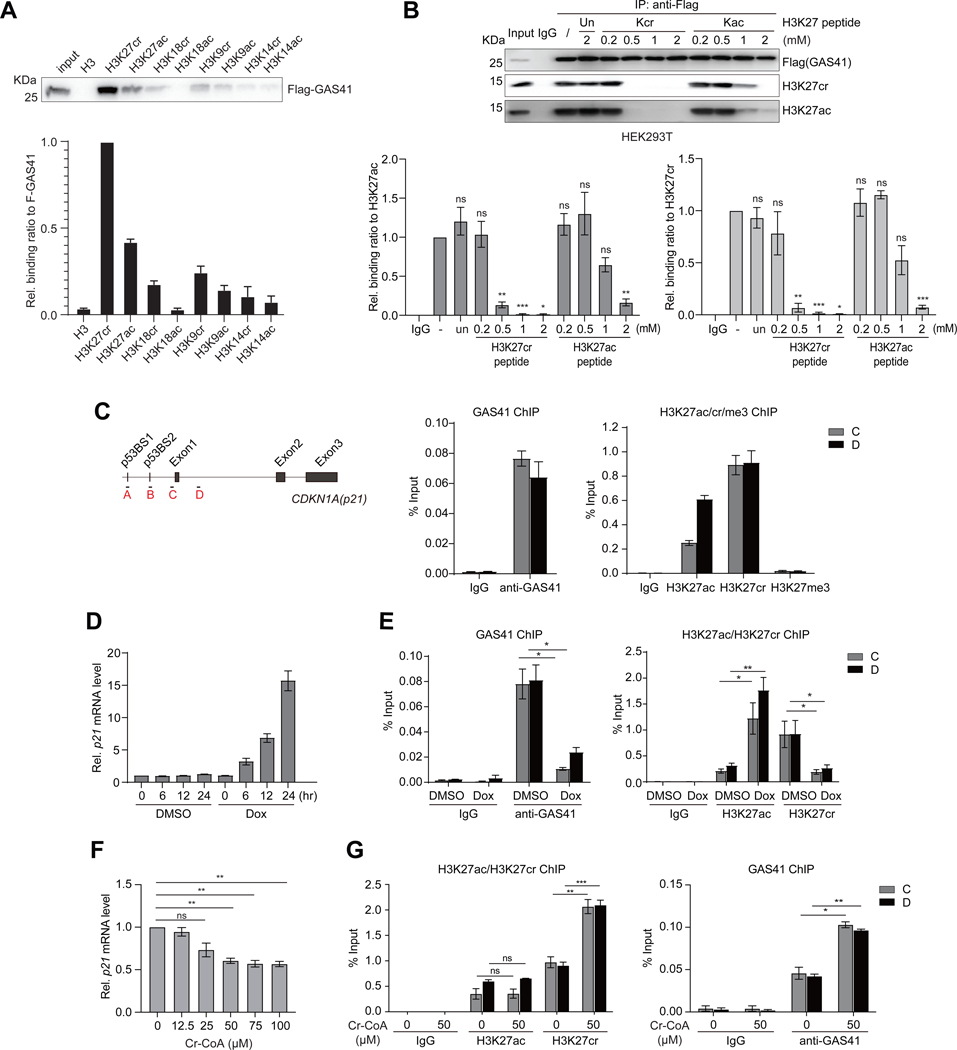
Figure 2. GAS41 regulates transcriptional repression of p21 mediated by H3K27cr.
(A) Western blots analysis and quantitation of ectopically expressed Flag-GAS41 in HCT116 cells binding to a set of biotinylated histone H3 peptides derived from H3 (residues 1–25, biotin-ARTKQTARKSTGGKAPRKQLATKAA), H3K27cr/ac (residues 21–33, biotin-ATKAAR-Kcr/Kac-S-APATG), H3K18cr/ac (residues 12–24, biotin-GGKAPR-Kcr/Kac-QLATKA), H3K9cr/ac (residues 3–15, biotin-TKQTAR-Kcr/Kac-SSGGKA), and H3K14cr/ac (residues 1–25, biotin-ARTKQTARKS-TGG-Kcr/Kac-APRKQLATKAA). n=3.
(B) Western blot analysis and quantitation of Flag-GAS41 binding to nucleosomal H3K27cr or H3K27ac in the lysate of HEK293T cells transiently transfected with Flag-GAS41 in the presence of unmodified histone H3K27, H3K27cr or H3K27ac peptide at concentrations as indicated. n=3.
(C) Scheme illustrating primer positions at p21 gene locus used in the study. Right, ChIP-qPCR analyses of endogenous GAS41(left), and histone H3K27cr, H3K27ac, or H3K27me3 (right) enrichment at p21 locus in HEK293T cells. n=3.
(D) mRNA transcript levels of p21 in HEK293T cells treated with Dox for 0, 6, 12, and 24 hours, respectively, n=3.
(E) ChIP-qPCR analyses showing changes of endogenous GAS41 (left), and H3K27ac or H3K27cr (right) enrichment at p21 locus in HEK293T cells treated with Dox for 24 hours, n=3.
(F) qPCR analysis of p21 expression in HEK293T cells treated with crotonyl-CoA at concentrations as indicated for 24 hours. n=3.
(G) ChIP-qPCR analysis of H3K27ac, H3K27cr (left), and endogenous GAS41 (right) enrichment at p21 locus in HEK293T cells with 24-hour crotonyl-CoA (50 μM) treatment. n=3.
We next sought to define how GAS41 targets p21 in chromatin. We detected enrichment of endogenous GAS41 and ectopic Flag-GAS41 at p21 locus in HEK293T cells by chromatin immunoprecipitation (ChIP) (Figures 2C and S3A). Both H3K27cr and H3K27ac are present at p21 locus in contrast to an extremely low level of H3K27me3 (Figures 2C and S2B). Consistent with its role in activating p21 transcription,34 transcription factor p53 was found markedly enriched at the p21 locus in HEK293T cells upon activation by DNA-damaging agent doxorubicin (Dox) that resulted in increased p53 Ser15-phosphorylation (Figures S3C and S3D). ChIP analysis revealed GAS41 displacement along with H3K27cr decrease and H3K27ac increase at the p21 locus upon p21 activation induced by Dox treatment in HEK293T and HCT116 cells (Figures 2D, 2E, and S3E-S3G). Similar effects on transcriptional de-repression of p21 were observed with p53 activation using Dox analog Idarubicin, or MDM2 inhibitor Nutlin3a in HEK293T and HCT116 cells (Figures S3H-S3K). Further, we observed a marked reduction of p21 mRNA level in HEK293T cells after 24-hour treatment of crotonyl-CoA (Figure 2F) that was accompanied with increased H3K27cr and GAS41 occupancy, but almost no change of H3K27ac at the p21 locus (Figure 2G). The latter agrees with the reported crotonyl-CoA stimulation of H3K18cr without significantly altering H3K18ac levels in cells.35 Collectively, these results indicate that GAS41 controls p21 transcriptional repression through its YEATS domain binding of H3K27cr.
GAS41 Works with MYC and SIN3A-HDAC1 Co-repressors for p21 Repression
To understand how GAS41 targets p21 in chromatin for transcriptional repression, we sought to determine its functional context that would require transcription factors and repressor proteins. Using reciprocal protein immunoprecipitation (IP) and western blotting analysis, we detected endogenous GAS41 association with MYC, and SIN3A-HDAC1,2 co-repressor complex proteins including SIN3A, HDAC1/2, RbAP48, and RBP1 in HEK293T cells (Figure 3A). IP study of Flag-GAS41 transfected HEK293T cells indicated GAS41 association with SIN3A-HDAC1,2 complex 36,37 to be selective, as no interaction was detected with NCoR, CHD3, or MBD2/3, key component proteins of the other known Mi-2/NuRD38 and NCoR39 co-repressor complexes (Figure S4A). The formation of GAS41/MYC/SIN3A-HDAC1 complex in cells was further supported by size exclusion chromatography analysis of HCT116 cell lysates (Figure S4B), and Flag-GAS41 co-localization with MYC, SIN3A, HDAC1, H3K27cr and H3K27ac at p21 locus observed in a sequential ChIP assay (Figure S4C). Note that Flag-GAS41 association with MYC/SIN3A-HDAC1 complex is likely to be functional as its expression level is similar to that of endogenous GAS41 and both are present in the complex as shown by co-IP (Figure S4D). Moreover, transfection of HEK293T cells with Flag-GAS41 and Flag-MYC reduced p21 protein level (Figure S4E), whereas knockdown of GAS41 or MYC increased p21 expression (Figure S4F), supporting the notion that GAS41 and MYC likely work together for p21 repression.
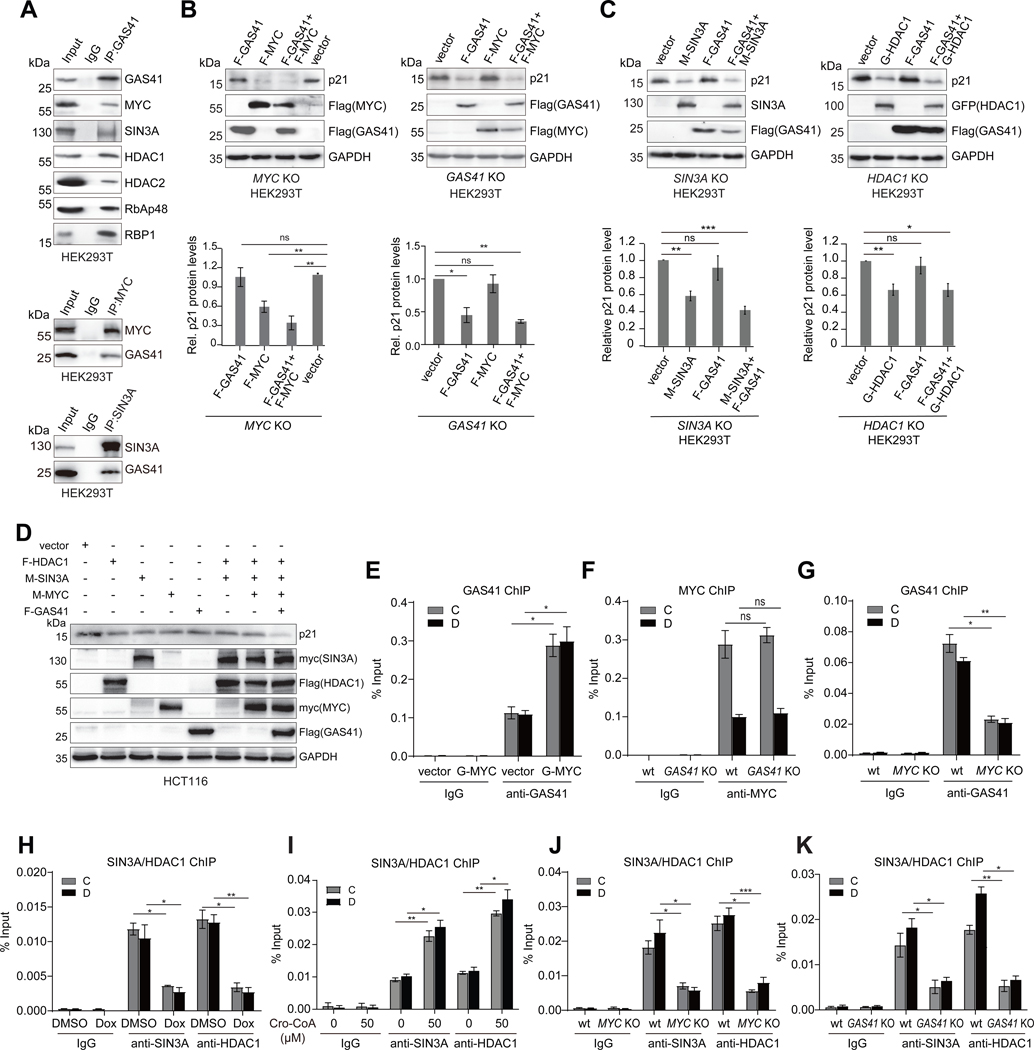
Figure 3. GAS41 works with MYC and SIN3A/HDAC1 co-repressors for p21 repression.
(A) Co-immunoprecipitation (Co-IP) of endogenous GAS41 and immunoblotting (IB) with specific antibodies assessing GAS41 association with MYC, SIN3A, HDAC1/2, RbAP48 and RBP1 in HEK293T cells (upper). Lower, reverse Co-IP showing endogenous MYC and SIN3A interactions with GAS41, respectively, in HEK293T cells.
(B) Western blot analysis and quantification of p21 expression in HEK293T MYC KO (left) and GAS41 KO (right) cells co-transfected with F-GAS41 and F-MYC, n=6 for MYC KO, n=3 for GAS41 KO.
(C) Western blot analysis and quantification of p21 expression in HEK293T SIN3A KO cells co-transfected with F-GAS41 and M-SIN3A (left), and HDAC1 KO cells co-transfected with F-GAS41 and G-HDAC1 (right). n=5 for SIN3A KO, and n=6 for HDAC1 KO.
(D) Western blot analysis of p21 expression in HCT116 cells co-transfected with F-HDAC1, M-SIN3A, M-MYC, and F-GAS41.
(E) ChIP-qPCR analysis of GAS41 enrichment at p21 locus in HEK293T cells transiently co-transfected with GFP empty vector, or GFP-MYC. n=3.
(F) ChIP-qPCR analysis of endogenous MYC enrichment at p21 locus in HEK293T wt and GAS41 KO cells. n=3.
(G) ChIP-qPCR analysis of GAS41 enrichment at p21 locus in HEK293T wt and MYC KO cells. n=3.
(H) ChIP-qPCR analysis of SIN3A and HDAC1 enrichment at p21 locus in HEK293T cells before and after 24-hour Dox treatment. n=3.
(I) ChIP-qPCR analysis of SIN3A/HDAC1 enrichment at p21 locus in HEK293T cells with 24-hour crotonyl-CoA (50 μM) treatment. n=3
(J) ChIP-qPCR analysis of SIN3A and HDAC1 enrichment at p21 locus in HEK293T wt and MYC KO cells. n=3.
(K)ChIP-qPCR analysis of SIN3A and HDAC enrichment at p21 locus in HEK293T wt and GAS41 KO cells. n=3. F-GAS41 stands for Flag-GAS41, F-MYC for Flag-MYC, F-HDAC1 for Flag-HDAC1, G-HDAC1 for GFP-HDAC1, M-SIN3A for myc-SIN3A, and M-MYC for myc-MYC.
To delineate the role of MYC/SIN3A-HDAC1 complex in GAS41 mediated p21 regulation, we generated HEK293T MYC, SIN3A, or HDAC1 KO cell lines using CRISPR/Cas9. Ablation of MYC, SIN3A, or HDAC1 all resulted in p21 induction, whereas in the CRISPR control cells, a three-base deletion in 5’ UTR (upstream of ATG) of MYC gene locus showed no effect on the expression of GAS41, MYC, HDAC1, SIN3A or p21 as compared to HEK293T cells (Figures S4G-S4J). Notably, co-transfection of Flag-GAS41 and Flag-MYC, not individually, was able to restore p21 repression in MYC or GAS41 KO cells, respectively (Figures 3B and S4K), confirming that both GAS41 and MYC are indispensable for p21 repression. Further, co-transfection of Flag-GAS41 with myc-SIN3A and GFP-HDAC1, respectively in HEK293T SIN3A or HDAC1 KO cells showed that SIN3A and HDAC1 were required for GAS41 function on p21 repression (Figure 3C). Our additional transfection studies of Flag-GAS41, Flag-MYC, GFP-HDAC1, or myc-SIN3A with HEK293T MYC, SIN3A, or HDAC1 KO cells established that p21 repression was compromised when any of these co-factors was missing (Figure S5A). Their functional interdependence was further confirmed by co-transfection of Flag-GAS41, myc-MYC, myc-SIN3A, and Flag-HDAC1 in HCT116 cells that yielded the most profound repression of p21 as compared to transfections lacking one or two factors (Figure 3D). Collectively, these results demonstrated the synergistic activities of GAS41 with MYC and SIN3A-HDAC1 co-repressors for transcriptional repression of p21.
Remarkably, Flag-GAS41 occupancy at p21 locus was significantly higher with its co-transfection with GFP-MYC than it alone (Figures 3E and S5B). MYC binding on p21 locus was not affected in GAS41 KO cells, whereas GAS41 binding on p21 locus was lost in MYC KO cells compared to wt cells (Figures 3F and 3G). Similarly, GAS41 association at p21 locus markedly decreased in MYC knockdown HCT116 cells (Figure S5C). Further, SIN3A and HDAC1 were found along with GAS41 at p21 locus, and their presence was reduced upon p21 de-repression after Dox, Ida, or Nutlin3a treatment (Figures 3H, S5D, S3F and S3H), or conversely increased upon p21 further repression with crotonyl-CoA treatment (Figure 3I). Importantly, SIN3A and HDAC1 occupancy at p21 locus was decreased in MYC or GAS41 KO cells (Figures 3J and 3K). These results confirmed that MYC is the transcription factor responsible for recruiting GAS41 to the p21 locus in chromatin, and that GAS41 and MYC work with SIN3A-HDAC1 co-repressor complex for p21 repression. Notably, this p21 transcriptional repression by MYC/GAS41/SIN3A-HDAC1 is different from the reported MYC/MIZ1 function for p21 suppression by displacing transcription co-activators at the p21 promoter,40,41 as we found that MIZ1 is not present in the GAS41/MYC complex (Figures S5E-S5I). These results suggest that the effect of MIZ1 on p21 transcription is likely influenced by its associating effector proteins.
GAS41 YEATS Domain Interacts with MYC and H3K27cr for p21 Repression
Despite being arguably one of the most recognized proto-oncogenes in tumorigenesis, the current mechanistic understanding of MYC function as transcription factor is largely limited to the DNA binding of its C-terminal basic/helix-loop-helix (bHLH) and leucine zipper (LZ) domains.42 To determine the molecular basis of the coordinated transcriptional activity of MYC and GAS41 in H3K27cr-mediated p21 repression, we mapped GAS41 and MYC interaction region using co-transfection of deletion constructs of Flag-GAS41 and GFP-MYC and IP assay. We found that the YEATS domain of GAS41 binds to the N-terminal MYC box II-III (MBII+III) region of MYC (residues 73–289) (Figures 4A, 4B, S6A and S6B). We validated this result in a hydrogen-deuterium exchange (HDX) study of MYC (residues 73–289) in the absence or presence of GAS41 YEATS domain (residues 11–150) followed by mass spectrometry (MS) analysis. The HDX-MS data showed that MYC residues 124–144, and to a lesser extent residues 260–266 exhibited a major reduction in the HDX rate in the presence of GAS41 as compared to the free MYC (Figures S6C and S6D, and Table S1), indicating that GAS41 YEATS domain binds primarily to MYC MBII and MBIIIb regions. Surprisingly, our NMR binding study revealed that MYC/GAS41 binding was competed off by a H3K27cr peptide, as MYC binding-induced line-broadening of amide resonances of 15N-GAS41 YEATS domain in the HSQC spectra was reversed upon addition of increasing amounts of H3K27cr peptide (Figures 4C and S6E).
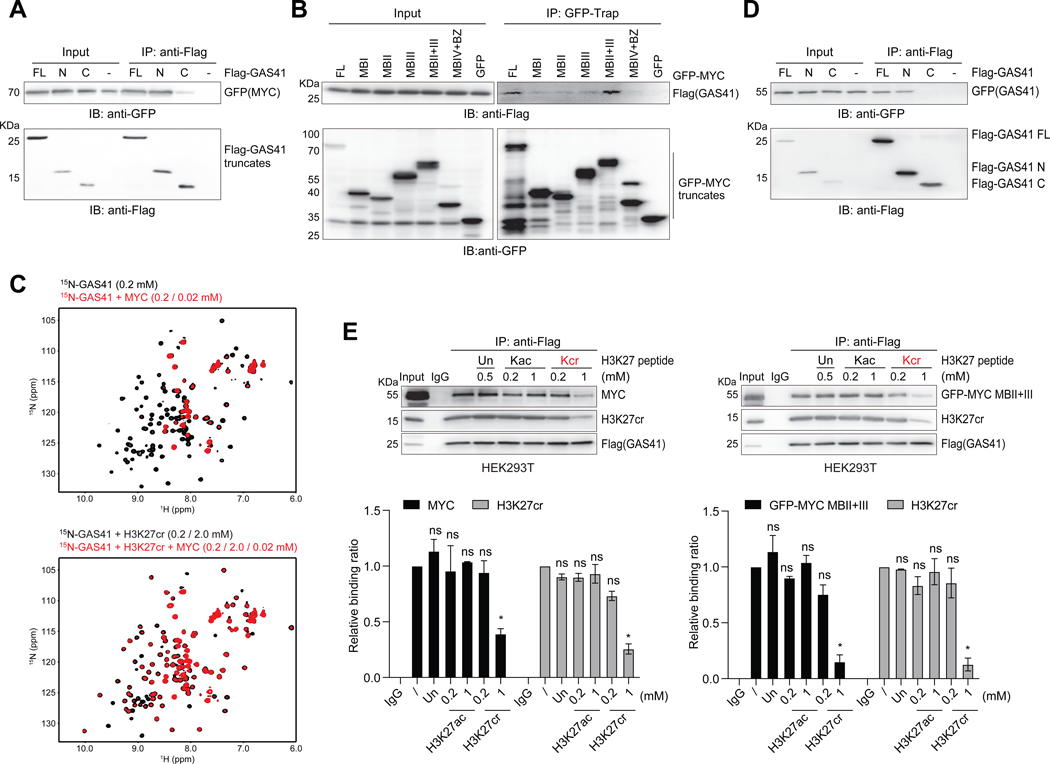
Figure 4. GAS41 YEATS domain interactions with H3K27cr and MYC.
(A) Co-IP of Flag-GAS41 full-length (FL; residues 1–227), N-terminal segment (N; residues 1–165) or C-terminal segment (C; residues 148–227) and IB with Flag and GFP antibodies assessing GAS41 interaction with MYC in HEK293T cells co-transfected with Flag-GAS41 and GFP-MYC.
(B) Co-IP of GFP-MYC and its various segments and IB with GFP and Flag antibodies in Flag-GAS41 and GFP-MYC co-transfected HEK293T cells.
(C) GAS41 YEATS domain binding to MYC (residues 1–262) (upper), or in the presence of the H3K27cr peptide (lower), as assessed by 2D 1H-15N HSQC spectra of the GAS41 YEATS domain.
(D) GAS41 dimerization, as assessed by Flag-GAS41 and GFP-GAS41 interaction in HEK293T cells, co-transfected with GFP-GAS41 and Flag-GAS41 of full-length, N- or C-terminal segment constructs, respectively. Cell lysate was subjected to Flag IP and followed by IB with antibodies against GFP.
(E) Western blot analysis and quantification of H3K27ac or H3K27cr peptide competition against Flag-GAS41 binding to MYC and H3K27cr in HEK293T cells. Varying amounts of H3K27, H3K27cr or H3K27ac peptide, as indicated, was added to the lysate of HEK293T cells, transfected with either Flag-GAS41 alone or together with GFP-MYC MBII+III, and followed by Flag IP and IB with antibodies against Flag, MYC, GFP, and H3K27cr. n=2.
To determine how GAS41 works with H3K27cr and MYC for p21 repression, we tested a notion of GAS41 dimerization and found that Flag-GAS41 was indeed co-immunoprecipitated with GFP-GAS41, co-transfected in HEK293T cells, and this interaction was mapped primarily to the N-terminal YEATS domain rather than the C-terminal coiled-coil domain of GAS41 (Figure 4D), even though the latter was suggested to contribute to GAS41 dimerization.43 Co-IP study showed that Flag-GAS41 is associated with H3K27cr and endogenous MYC or GFP-MYC MBII+III in HEK293T cells, and a H3K27cr peptide, but not a H3K27ac peptide at the same concentrations, can dissociate this MYC/Flag-GAS41/H3K27cr complex (Figure 4E). The latter highlights the specificity of GAS41 YEATS domain binding to H3K27cr over H3K27ac in a cellular context. From these results, we concluded that GAS41 likely through its N-terminal YEATS domain forms a dimer, and that the YEATS domain of each protomer interacts with H3K27cr or MYC, thereby forming the MYC/GAS41/H3K27cr complex that works with the SIN3A-HDAC1 co-repressors for p21 transcriptional repression in chromatin.
Structural Basis of GAS41 YEATS Domain Recognition of H3K27cr and MYC
We next sought to determine the detailed molecular basis of GAS41 YEATS domain recognition of H3K27cr and MYC through protein structural analysis. However, the highly dynamic nature of protein-protein interactions prevented us from determining 3D structure of GAS41 YEATS domain in complex with MYC. Instead, we solved a 2.30Å resolution crystal structure of the complex of GAS41 YEATS domain bound to a H3K27cr peptide (Figures 5A and S7A, and Table S2). This crystal contains two molecules (A and B) per asymmetric unit, and their overall structures are nearly the same with a root-mean-square deviation of 0.75Å for the Cα atoms excluding a short helix (α2, residues 123–128) (Figures S7B and S7C). This short helix is missing in nearly all reported structures of the GAS41 YEATS domain solved in different forms possibly due to the high structural flexibility of this region.30 We noticed the possibility that the GAS41 YEATS domain forms a tetramer structure as a dimer of dimer that is related by crystallographic 2-fold symmetry axis (Figure S7B). We obtained experimental evidence in solution for only a dimeric form, but not a tetrameric form of the GAS41 YEATS domain. Future study is needed to investigate GAS41 oligomeric state in a functional context.
Figure 5. Molecular basis of GAS41 YEATS domain interactions with H3K27cr and MYC for p21 repression.
(A) Crystal structure of GAS41 YEATS domain (blue) in complex with H3K27cr peptide (yellow) as shown in a dimer (molecules A and B). Right, Electrostatic potential surface representation of GAS41 YEATS domain bound to H3K27cr peptide (upper), and the detailed structural basis of GAS41 YEATS domain recognition of the H3K27cr peptide (lower), as depicted in molecule A. Side chains of key residues engaged in protein/peptide interactions are color-coded by atom type, and a hydrogen bond is indicated by a dashed line (red).
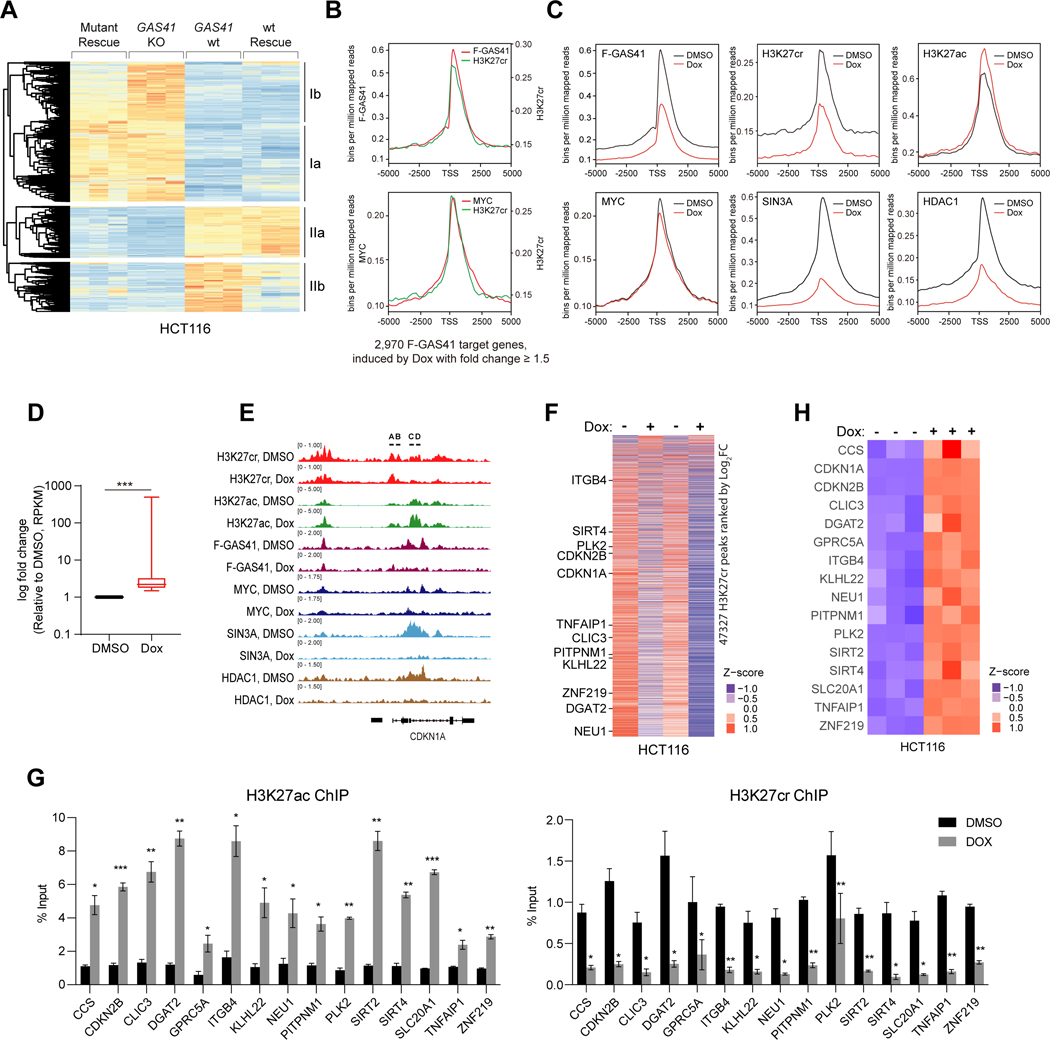
(B) Effects of mutations of acyl-lysine binding residues on GAS41 binding to H3K27cr, MYC and SIN3A-HDAC co-repressor complex proteins. The lysate of HEK293T cells transfected with Flag-GAS41 wt or mutants was subjected to Flag IP and followed by IB with antibodies against Flag, H3K27cr, MYC, SIN3A and HDAC1, respectively. n=2.
(C) Effects of mutations of acyl-lysine binding residues in GAS41 on p21 expression in HEK293T cells transfected Flag-GAS41 wt or corresponding mutants. Quantification is shown as indicated. n=3.
(D) Western blot analysis of p21 expression in HCT116 GAS41 KO cells stably expressing Flag-GAS41 wt, Y74A/W93A mutant, or empty vector.
(E, F) Effects of H43S mutation on GAS41 binding to H3K27ac, H3K27cr and MYC. The lysate of HCT116 cells transfected with Flag-GAS41 wt and H43S was subjected to Flag IP and followed by IB with antibodies against Flag, H3K27ac, H3K27cr and MYC respectively. n=3.
(G) Western blot analysis of p21 expression in HCT116 expressing Flag-GAS41 wt, H43S mutant, or empty vector. n=4.
(H) ChIP-qPCR analysis showing Flag-GAS41 occupancy at p21 locus in HCT116 cells expressing Flag-GAS41 wt and H43S, n=3.
Our crystal structure shows that crotonyl-lysine is engaged in a π–π–π sandwich binding mechanism established between H3K27cr and aromatic side chains of His43, Tyr74 and Trp93 (Figure 5A), similar to the AF9 6,32 or Taf14 44 YEATS domain recognition of H3K9cr and H3K18cr. The crotonyl-lysine recognition by GAS41 is bolstered by a 2.7Å hydrogen bond (molecule A) formed between side-chain hydroxyl oxygen of Ser73 and amide nitrogen of the crotonyl-lysine. While it can also form a hydrogen bond to Ser73, an acetyl-lysine lacks the electron-rich double bond moiety of crotonyl-lysine, and likely has reduced aromatic and hydrophobic interactions with Tyr74 and Trp93. Further, because of its shorter side chain than that of crotonyl-lysine, acetyl-lysine may not interact with the imidazole of His43 located at the far end of the aromatic cage, explaining lower affinity of H3K27ac than H3K27cr to the GAS41 YEATS domain.30 Our structure also illuminated the selectivity of the GAS41 YEATS domain for H3K27cr over H3K9cr or H3K18cr, which is likely due to recognition of Pro30 in the H3K27cr peptide by a small hydrophobic/aromatic concave surface formed by Leu120 and Phe121 (Figure 5A). The latter Phe121 is unique in GAS41 and not conserved among the YEATS domain proteins (Figure S7A).
To define MYC binding site in GAS41, we obtained NMR backbone resonance assignment of its YEATS domain using 3D triple-resonance NMR methods.45 Based on MYC binding-induced NMR resonance perturbation of the YEATS domain, we mapped MYC binding in GAS41 to the H3K27cr binding region (Figure S8A). The MYC binding site of GAS41 was confirmed by IP and mutation analysis of H3K27cr-binding residues Tyr74 and Trp93 whose substitution to Ala, alone or in combination, markedly reduced GAS41 binding to MYC, and almost completely abolished binding to H3K27cr, SIN3A and HDAC1 (Figure 5B). The Y74A/W93A mutation did not compromise the YEATS domain structure, as shown by the NMR HSQC spectra (Figure S8B). Single or double mutation of Y74A and W93A in Flag-GAS41 diminished its ability for p21 repression in HEK293T cells (Figure 5C). To fully determine the importance of GAS41/H3K27cr binding for p21 repression in cells, we established the wt and Y74A/W93A mutant GFP-Flag-GAS41 stable cell lines from HCT116 GAS41 KO cells (Figure S8C). As compared to the GAS41 wt, Y74A/W93A mutant was not able to reinstall p21 repression in HCT116 GAS41 KO cells, thus failed to restore the cell growth (Figures 5D and S8D), and showed much lower occupancy at the p21 promoter (Figure S8E), confirming the critical role of GAS41/H3K27cr binding for GAS41 association with chromatin. To determine the function of H3K27cr-binding deficient mutant with expression at the physiological level, we established GAS41 Y74A heterozygous HEK293T cell line with CRISPR/Cas9 (Figure S8F), of which GAS41 alleles are either Y74A mutant or knockout. These Y74A mutant cells lost p21 repression and exhibited decreased cell growth rate and impaired cell cycle progression (Figure S8G).
To distinguish GAS41 binding to H3K27cr vs. H3K27ac in MYC/GAS41/SIN3A-HDAC1 complex-directed gene transcriptional repression in chromatin, we sought to identify a GAS41 mutant that would abrogate H3K27cr binding without affecting H3K27ac binding. We focused on His43 because its mutation to Ala exhibited a small but statistically significant reduction of GAS41 binding to H3K27cr and p21 repression, yet showed nearly full H3K27ac binding (Figures 5B, 5C, S8H and S8I). We carried out a holistic site-directed mutagenesis study by changing His43 to 18 other amino acids and examined these GAS41 mutants in binding to biotin-tagged H3K27, H3K27ac and H3K27cr peptides using ELISA (data not shown). From this study, we discovered that GAS41 H43S mutant shows over 60% reduction in H3K27cr binding but retains nearly intact H3K27ac binding as demonstrated by Co-IP assay of cell extracts of HCT116 cells transiently transfected with Flag-GAS41 wt or H43S mutant (Figure 5E). This agrees with binding affinity measurements in a microscale thermophoresis assay, which showed that GAS41-H43S exhibited 6-fold reduction in binding to H3K27cr vs. about 2-fold reduction to H3K27ac as compared to GAS41 wt (Figure S8J). GAS41-H43S retains MYC binding and its transfection does not affect H3K27ac abundance, but nearly completely loses its ability to bind chromatin at the p21 locus, which is accompanied with transcriptional de-repression of p21 in HCT116 cells (Figures 5F–5H and S8K). From these results, we concluded that GAS41 occupancy on chromatin is likely dependent on its binding to H3K27cr, but not H3K27ac, and that the YEATS domain/H3K27cr binding is essential for GAS41 to repress p21 expression in chromatin.
H3K27cr is a Distinct Histone Mark for Gene Transcriptional Repression
To elucidate a genome-wide role of H3K27cr in gene transcriptional repression by the GAS41/MYC/SIN3A-HDAC1 co-repressor complex, we performed RNA-seq study of HCT116 GAS41 wt and GAS41 KO cells, as well as GAS41 KO cells that stably expressed GFP-Flag-GAS41 wt (wt rescue), or GFP-Flag-GAS41-Y74A/W93A (mutant rescue) (Figure 6A). Principal component analysis (PCA) showed excellent agreement between independently prepared triplicate samples (Figure S9A). GAS41 knockout resulted in up-regulation of 1674 genes and down-regulation of 961 genes (Table S3). Our hierarchical clustering analysis allowed us to segregate genes into two categories, i.e. genes dependent upon or independent of the acyl-lysine binding activity of the GAS41 YEATS domain (Figures 6A and S9B). Genes in each category are further grouped into two clusters (Clusters Ia & Ib vs. IIa & IIb) owing to their degree of the YEATS domain dependency, i.e. nearly full or partial dependence.
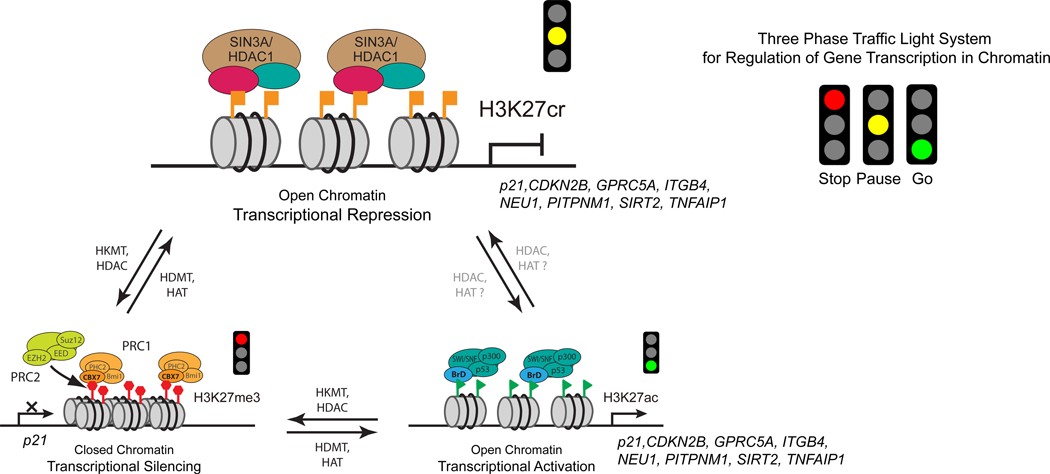
Figure 6. H3K27cr regulates gene transcriptional repression in chromatin.
(A) Heatmap of RNA-seq counts of HCT116 GAS41 wt, GAS41 KO, wt rescue, and Y74A/W93A mutant rescue cells. Genes selected are significantly changed (P <0.1) and ranked by log2[fold change] >=1.5 between wild-type and GAS41 KO samples. Counts are represented as z-scores between all samples for each gene. Genes responded to wt or mutant rescue are grouped into 2 categories whose transcription is either fully or partially repressed by (Ia & Ib), or positively dependent fully or partially upon GAS41 (IIa & IIb).
(B) ChIP-seq peaks of H3K27cr vs. Flag-GAS41 (upper), and H3K27cr vs. MYC (lower) (average of 2–3 samples), centered at the TSS (±5 kb) of 2,970 GAS41 target genes in HCT116 cells that showed transcriptional induction upon Dox treatment with fold change ≥ 1.5. Reads were normalized to bins per million mapped reads.
(C) ChIP-seq peaks of Flag-GAS41, MYC, SIN3A, HDAC1, H3K27cr and H3K27ac (average of 2–3 samples), centered at the TSS (±5 kb) of the 2,970 GAS41 target genes in HCT116 cells before and after Dox treatment. Reads were normalized to bins per million mapped reads.
(D) Box-and-whisker plot (min to max) of RNA-seq data showing fold change of all genes that are stimulated upon Dox treatment in HCT116 cells and bound by Flag-GAS41 within ±5 kb TSS (total 2,970 genes). *** p<0.001.
(E) Genome browser view of H3K27cr, H3K27ac, Flag-GAS41, MYC, SIN3A and HDAC1 ChIP-seq peaks at the p21(CDKN1A) locus in DMSO and Dox treated HCT116 cells.
(F) Heatmap of H3K27cr peak intensities at H3K27cr peaks as identified by ChIP-seq in DMSO and Dox treated HCT116 cells, with each replicate sample represented as one column.
(G) ChIP-qPCR analysis of H3K27ac and H3K27cr occupancy at the gene loci of CCS, CDKN2B, CLIC3, DGAT2, GPRC5A, ITGB4, KLHL22, NEU1, PITPNM1, PLK2, SIRT2, SIRT4, SLC20A1, TNFAIP1 and ZNF219 in HCT116 cells before and after Dox treatment. Values are normalized to DMSO control group. n=3.
(H) Heatmap of RNA-seq datasets of triplicate samples of HCT116 cells showing selected genes that fulfill the following criteria: differentially expressed in GAS41 KO cells, targeted by GAS41 as identified by ChIP-seq, and showed decreased H3K27cr occupancy upon Dox treatment as shown by ChIP-seq.
See also Figure S9, Figure S10 and Table S3.
We next defined chromatin occupancy of GAS41, MYC, SIN3A, HDAC1, H3K27cr, and H3K27ac in HCT116 cells in a ChIP-seq study. While the GAS41 antibody works for ChIP-qPCR, it did not produce quality ChIP-seq data. Accordingly, we performed ChIP-seq using Flag-GAS41 transfected in HCT116 cells, in which Flag-GAS41 expression was comparable to endogenous GAS41, and both Flag-GAS41 and endogenous GAS41 are present in the MYC/SIN3A-HDAC1 complex (see Figure S4D). Concordant with our finding that GAS41 functions in association with MYC and SIN3A-HDAC1 co-repressors, genomic occupancy of Flag-GAS41, MYC, SIN3A and HDAC1 appears correlated (Figure S9C). We found that 86.0% of Flag-GAS41-bound genes were occupied by MYC or SIN3A-HDAC1, and 43.6% of Flag-GAS41-bound genes were co-occupied by MYC and SIN3A-HDAC1 (Figure S9D), while 67.1% of Flag-GAS41 genes were overlapped with H3K27cr (Figure S10E). Flag-GAS41 occupancy at transcription start sites (TSS) of 12,862 GAS41 target genes showed strong enrichment for both H3K27cr and MYC (Figure S9F). A consensus DNA binding analysis using the Homer program revealed that GAS41 and MYC share common binding motifs including MYC (CACGTG) (Figure S9G). Of these GAS41 target genes, 2,970 genes were co-occupied by MYC and SIN3A-HDAC1, and showed transcriptional up-regulation by ≥1.5-fold upon Dox treatment in HCT116 cells, which was correlated to increased levels of H3K27ac and decreased levels of H3K27cr, Flag-GAS41, SIN3A, HDAC1, and to a lesser extent MYC at the TSS (Figures 6B–6D). Collectively, these results support the notion that GAS41, MYC, SIN3A, HDAC1 and H3K27cr work together to regulate gene transcription.
Our ChIP-seq analysis showed clearly reduced occupancy of Flag-GAS41, MYC, SIN3A, HDAC1, and H3K27cr, and conversely, increased presence of H3K27ac along the p21(CDKN1A) locus after Dox treatment (Figure 6E). Notably, H3K27cr levels exhibited a global reduction in HCT116 cells after Dox treatment (Figure 6F). GAS41 target genes appear to be involved a diverse set of key cellular processes for cell maintenance and proliferation including cell cycle, cellular senescence, p53 signaling pathway, DNA replication, and colorectal cancer, as shown by KEGG enrichment analysis of our genomic sequencing data (Figure S9H). Like p21, after Dox treatment, an increase of H3K27ac, and a decrease of H3K27cr, GAS41 and MYC presence in chromatin were consistently seen for a selected group of GAS41 target genes (Cluster Ia) including CCS, CDKN2B, CLIC3, DGAT2, GPRC5A, ITGB4, KLHL22, NEU1, PITPNM1, PLK2, SIRT2, SIRT4, SLC20A1, TNFAIP1 and ZNF219 (Figures 6G, S9I, and S9J). These genes showed an increase in mRNA transcripts in HCT116 cells after Dox treatment, both in the RNA-seq data and individual validation by qPCR (Figures 6H and S9K). Additionally, crotonyl-CoA treatment caused transcriptional repression for most of these selected GAS41 target genes in HEK293T cells, which were accompanied with increased H3K27cr and unchanged H3K27ac levels (Figures S9L and S9M). Furthermore, a global analysis of HCT116 GAS41 KO cells stably expressing Flag-GAS41 or Flag-GAS41-H43S revealed that H3K27cr-binding deficient H43S mutation caused a major decrease of GAS41 occupancy in chromatin (Figures S10A-S10C). This decrease of chromatin binding of GAS41-H43S was corroborated with this GAS41 mutant’s reduced ability for gene transcriptional repression, as shown with the same set of selected GAS41 target genes (Figure S10D). Taken together, our genomic study confirmed that H3K27cr likely represents a distinct histone mark for gene transcriptional repression exerted by a novel GAS41/MYC/SIN3A-HDAC1 co-repressor complex in chromatin.
GAS41 has been report to function in gene transcriptional activation and repression in cells13,46–49 that are likely influenced by its associating proteins. Therefore, it is prudent to examine the experimental evidence in a biological context. For instance, Shi et al. reported that GAS41 works with TIP60 acetyltransferase complex to facilitate H2A.Z deposition in transcriptional activation, and GAS41 or TIP60 knockdown leads to reduced H2A.Z abundance and expression of cell cycle genes30. We observed that GAS41 is associated with the TIP60 complex in HCT116 cells, but GAS41 KO does not affect TIP60 association with the other components of the complex (Figure S10E). TIP60 binding at GAS41 target genes in GAS41 KO cells is higher than that in wild type cells and is important for transcriptional expression of GAS41 target genes including p21 (Figures S10F and S10G), suggesting TIP60 acts in transcriptional activation after GAS41 dissociation from chromatin. Notably, H2A.Z occupancy at GAS41 target genes is lower in HCT116 GAS41 KO cells than wild type cells (Figure S10H), but similar in GAS41 KO cells stably expressing Flag-GAS41 wt and H43S (Figure S10I). Therefore, our results indicate that GAS41 function in gene transcriptional repression, in association with the MYC/SIN3A-HDAC1 corepressor complex and mediated by H3K27cr, is independent of the TIP60 complex for transcriptional activation, and also different from the GAS41 function with TIP60 for H2A.Z deposition. The latter likely does not involve GAS41 YEATS domain binding to H3K27cr.
DISCUSSION
Histone H3K27me3 and H3K27ac have long been recognized as two chromatin states marked for long-term gene transcriptional silencing in compact heterochromatin and gene transcriptional activation with enhancer elements in open chromatin, respectively. Here we report that H3K27cr represents a previously unrecognized, distinct chromatin state for gene transcriptional repression (Figure 7). This H3K27cr-directed transcriptional repression operates through selective recognition of H3K27cr by the YEATS domain of GAS41 that is recruited by transcription factor MYC to target genes in chromatin and works in concert with the SIN3A-HDAC1 co-repressors, as demonstrated for cell cycle inhibitor p21 and confirmed with many target genes. This H3K27cr is different from the “bivalent chromatin”, marked by H3K4me3 and H3K27me3, as the role of H3K27cr in gene regulation is defined with a chemically distinct histone modification and molecular recognition by a structurally distinctive histone reader domain. The latter feature is on a par with H3K27ac recognition by the bromodomain2 and H3K27me3 by the chromodomain.50
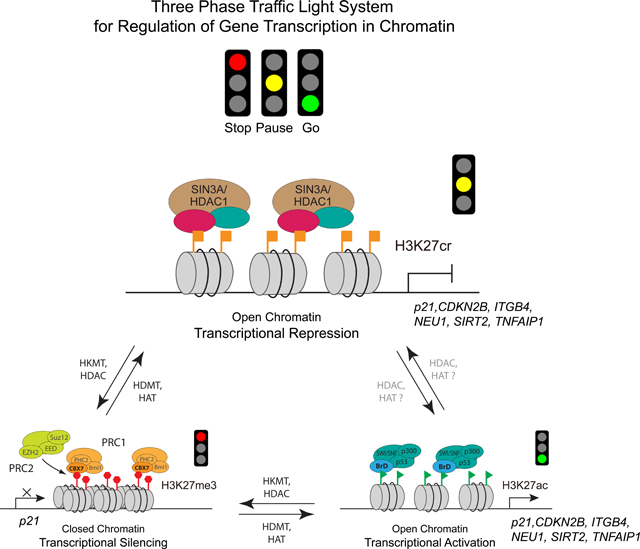
Figure 7. H3K27cr represents a distinct chromatin state for gene transcriptional repression.
Schematic illustration of a “Three-phase traffic light system” model describing three different H3K27 modifications as distinct chromatin states for gene transcription, i.e. H3K27me3 (Stop) marks for transcriptional silencing, H3K27cr (Pause) for transcriptional repression, and H3K27ac for transcriptional activation (Go). As illustrated for p21 and other GAS41 target genes such as those validated in Figure 6H, H3K27cr-directed gene transcriptional repression operates through selective recognition of H3K27cr by the YEATS domain of GAS41 in complex with MYC and SIN3A/HDAC1 co-repressor complex in chromatin.
We propose a model of “Three-phase traffic light system” to describe the role of histone modifications at H3K27 in regulation of gene transcription in chromatin in that H3K27me3 (Stop), H3K27cr (Pause), and H3K27ac (Go) symbolize three functionally distinct histone modification states for gene transcriptional silencing, repression and activation, respectively (Figure 7). This three-phase switch mechanism offers quick yet ordered coordination between chromatin structure and gene transcription, providing long-sought evidence in support of the postulation that different modifications on one single histone residue can dictate different outcome of gene transcription.51,52 Regulation of histone modifying enzymes and modification-specific histone readers in association with these three chromatin states endows histones with the capacity to direct on-demand gene expression or repression in response to physiological and environmental cues. Mis-regulation of these distinct chromatin states can lead to disease development. In this context, our study provides a mechanistic explanation for the previously reported association of MYC with GAS41 22,40,53–58 for negative regulation of p21 expression and explains our observed gene amplification of GAS41 and MYC in colorectal cancer tissues that is inversely correlated to the downregulation of p21 (Figure 1B).
While this H3K27cr-mediated gene transcriptional repression mechanism operates likely under certain functional conditions in gene regulation or mis-regulation in diseases, understanding of the details about the control of histone lysine acylation modifying enzymes and how they direct transition between H3K27cr and H3K27ac states warrants further investigation. Our study opens a door for developing new anti-cancer therapy that works through pharmacological induction of transcriptional de-repression of tumor suppressor genes often over-repressed in human cancers including colorectal cancer by H3K27cr-mediated GAS41 and MYC functions. The latter is arguably one of the most recognized proto-oncogenes in tumorigenesis, but its mechanism as transcription factor for gene repression has remained very limited.59 Finally, our study illuminates a new dimension of the sophisticated histone biology in gene regulation in association in part with these newly discovered distinct histone modifications,1,51 and highlights the importance of uncovering their functional mechanisms in human biology and diseases.
Limitations of the Study
While we showed that MYC directly binds the GAS41 YEAST domain, we do not know the detailed molecular basis of MYC and GAS41 interaction. Further, while our results suggest that GAS41 YEATS domain likely forms a dimer, which enables GAS41 to interact with both H3K27cr and MYC for GAS41 function in gene transcriptional repression in cells, we do not know how GAS41 simultaneously interacts with H3K27cr and MYC at the target gene loci in chromatin. We expect that GAS41 function in control of gene transcriptional activation and repression through its YEATS domain binding to histone lysine crotonylation or acetylation is to be context dependent, but a better understanding warrants further investigation. Also, our study of GAS41 KO cell rescue with GAS41 wild-type or acyl-lysine deficient mutant suggests GAS41 functions in gene transcription beyond the acyl-lysine binding, which requires further study. Finally, it is known that histone lysine crotonylation level in cells is normally much lower than that of histone lysine acetylation, but their levels can change under special circumstance such as cellular metabolism.12,35 Therefore, the role of H3K27cr in regulation of gene transcriptional repression shall be further examined under different biological conditions.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Ming-Ming Zhou (ming-ming.zhou@mssm.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
All structure coordinates were deposited in the Protein Data Bank (PDB) with identification number 8I60. Genomic sequencing datasets from this study were deposited to the NCBI Gene Expression Omnibus (GEO) data repository under accession number GSE128590. Source image files for figures are deposited to Mendeley Data. All data are publicly available as of the date of publication. Accession numbers and DOI are listed in the Key Resources Table.
This paper does not report any original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-H3K27ac Antibody | Abcam | Cat# ab4729, RRID:AB_2118291 |
| Rabbit anti-H3K27ac Antibody | Millipore | Cat# 07-360, RRID: AB_310550 |
| Rabbit anti-H3K27cr Antibody | ThermoFisher Scientific | Cat# 712478, RRID:AB_2848247 |
| Rabbit anti-H3 Antibody | Abcam | Cat# ab1791, RRID:AB_302613 |
| Rabbit anti-RbAp48 Antibody | Abcam | Cat# ab79416, RRID:AB_1603754 |
| Rabbit anti-MYC Antibody | Abcam | Cat# ab32072, RRID:AB_731658 |
| Mouse anti-MYC Antibody | Santa Cruz | Cat# sc-40, RRID:AB_627268 |
| Rabbit anti-NCoR Antibody | Abcam | Cat# ab24552, RRID:AB_2149005 |
| Rabbit anti-MBD2 Antibody | Abcam | Cat#ab188474 |
| Rabbit anti-MBD3 Antibody | Abcam | Cat#ab157464 |
| Rabbit anti-CHD3 Antibody | Abcam | Cat# ab109195, RRID:AB_10862514 |
| Rabbit anti-p21 Antibody | Abcam | Cat# ab109199, RRID:AB_10861551 |
| Rabbit anti-GAPDH Antibody | Abcam | Cat# ab9485, RRID:AB_307275 |
| Rabbit anti-RBP1 Antibody | Abcam | Cat# ab154881, RRID:AB_2721835 |
| Mouse anti-H3K27cr Antibody | PTM Biolabs | Cat#PTM-Bio 526 |
| Mouse anti-GFP Antibody | CMCTAG | Cat#AT0028 |
| Rabbit anti-GFP Antibody | CMCTAG | Cat#AT0044 |
| Mouse anti-Flag Antibody | CMCTAG | Cat#AT0022 |
| Rabbit anti-Flag Antibody | CMCTAG | Cat#AT0502 |
| Rabbit anti-c-Myc tag Antibody | CMCTAG | Cat#AT0045 |
| Mouse anti-c-Myc tag Antibody | CMCTAG | Cat#AT0023 |
| Mouse anti 6xHis-tag Antibody | BBI Life Sciences | Cat#D191001 |
| Rabbit anti-SIN3A Antibody | Cell Signaling Technologies | Cat# 8056, RRID:AB_11217624 |
| Rabbit anti-H3K27me3 Antibody | Cell Signaling Technologies | Cat# 9733, RRID:AB_2616029 |
| Normal rabbit IgG | Cell Signaling Technologies | Cat# 2729, RRID:AB_1031062 |
| Normal mouse IgG | Santa Cruz | Cat# sc-2025, RRID:AB_737182 |
| Rabbitanti-HDAC1 Antibody | Active motif | Cat# 40967, RRID:AB_2614948 |
| Rabbit anti-HDAC2 Antibody | Proteintech | Cat# 16152-1-AP, RRID:AB_2118514 |
| Mouse anti-alpha-tubulin Antibody | Proteintech | Cat# 66031-1-Ig, RRID:AB_11042766 |
| Mouseanti-GAS41 Antibody | Santa Cruz | Cat# sc-393708, RRID:AB_2892567 |
| Rabbit anti-GAS41 Antibody | Genetex | Cat# GTX16452, RRID:AB_2887538 |
| Mouseanti-Actin Antibody | Beyotime | Cat# AA128, RRID:AB_2861213 |
| Goat anti-Mouse IgG H&L (HRP) 2ND Antibody | Abcam | Cat# ab97040, RRID:AB_10698223 |
| Goat anti-Rabbit IgG H&L (HRP) 2ND Antibody | Abcam | Cat# ab97080, RRID:AB_10679808 |
| Mouse anti-FlagAntibody | Sigma | Cat# F1804, RRID:AB_262044 |
| Rabbit anti H2A.Z Antibody | Cell Signaling Technologies | Cat# 50722, RRID:AB_2799379 |
| Rabbit anti TIP60 Antibody | Protein tech | Cat# 10827-1-AP, RRID:AB_2128431 |
| Bacterial and virus strains | ||
| E. coli BL21(DE3) | TransGen Biotech | Cat# CD601 |
| E. coli DH5alpha | TransGen Biotech | Cat# CD201-02 |
| E. coli RFzero | Mukai et al. (2011)60 | N/A |
| E. coli JM109 (DE3) | Promega | P980A |
| Biological Samples | ||
| Human colorectal cancer cDNA microarray | OutdoBiotech | Cat# cDNA-HColA060CS02 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Protease inhibitor Cocktail | Roche | Cat# 11873580001 |
| Phusion High-Fidelity DNA Polymerase | ThermoFisher Scientific | Cat# F530S |
| RNAiMAX Transfection Reagent | Invitrogen | Cat# 13778150 |
| TRIzol reagent | Invitrogen | Cat# 15596026 |
| Doxorubicin | Selleck | Cat# S1208 |
| Idarubicin | Selleck | Cat# S1228 |
| Nutlin-3a | Selleck | Cat# S8059 |
| Blasticidin S HCl | Selleck | Cat# S7419 |
| Bezonase Nuclease | Sigma | Cat# E1014-25KU |
| Crotonyl-CoA | Sigma | Cat. 28007, lot BCBZ4793 |
| Dynabeads® Protein G | Invitrogen | Cat# 10004D |
| Straptavidin Magnetic beads | Engibody | Cat# IF9042 |
| GFP-Trap | ChromoTek | Cat# GTM-20 |
| H-Lys(Ac)-OH | BACHEM | Cat# 4015284 |
| H-Lys(crotonyl)-OH | BLDpharm | Cat# BD328520 |
| Synthesized H3 K27cr (a.a. 21-33) peptide (ATKAAR-Kcr-SAPATG) | This paper | Mimotopies |
| Synthesized H3 K27ac (a.a. 21-33) peptide (ATKAAR-Kac-SAPATG) | This paper | Mimotopies |
| Synthesized H3 K27 (a.a. 21-33) peptide (ATKAARKSAPATG) | This paper | Mimotopies |
| Synthesized H3 K9cr (a.a. 3-15) peptide (TKQTAR-Kcr-STGGKA) | This paper | Mimotopies |
| Synthesized H3 K9ac (a.a. 3-15) peptide (TKQTAR-Kac-STGGKA) | This paper | Mimotopies |
| Synthesized H3 K18cr (a.a. 12-24) peptide (GGKAPR-Kcr-QLATKA) | This paper | Mimotopies |
| Synthesized H3 K18ac (a.a. 12-24) peptide (GGKAPR-Kac-QLATKA) | This paper | Mimotopies |
| Synthesized H3 (a.a.1-25) peptide (ARTKQTARKSTGGKAPRKQLATKAA) | This paper | Mimotopies |
| Synthesized H3 K14cr (a.a.1-25) peptide (ARTKQTARKSTGG-Kcr-APRKQLATKAA) | This paper | Mimotopies |
| Synthesized H3 K14ac (a.a.1-25) peptide (ARTKQTARKSTGG-Kac-APRKQLATKAA) | This paper | Mimotopies |
| Recombinant protein YEATS4 (a.a. 11-150) | This paper | N/A |
| Recombinant protein YEATS4 (a.a. 11-165) | This paper | N/A |
| Recombinant protein YETAS4 (a.a. 11-150) Y74A/W93A | This paper | N/A |
| Recombinant protein c-Myc (a.a. 1-262) | This paper | N/A |
| Recombinant protein mGFP YEATS4 | This paper | N/A |
| Critical Commercial Assays | ||
| Re-ChIP-IT | Active Motif | Cat# 53016 |
| Streptavidin-coated 96-well plate | Thermo Scientific | Cat# 15501 |
| QuickChange Site-Directed Mutagenesis Kit | Agilent Technologies | Cat# 200518 |
| Power SYBR green PCR master mix | Applied Biosystems | Cat# A25778 |
| EasyScript One-Step gDNA Removal and cDNA Synthesis SuperMix | Transgen | Cat# AE311-03 |
| Plasmid Plus Midi Kit | Qiagen | Cat# 12943 |
| Deposited Data | ||
| GAS41 YEATS domain/H3K27cr structure | This paper | PDB:8I60 |
| RNA-seq and ChIP-seq data | This paper | GSE128590 |
| Western blot and cell fluorescence images | This paper | Mendeley Data: https://dol.org/10.17632/wn5dgnz64x.1 |
| Experimental Models: Cell Lines | ||
| HEK293T | COBIOER | Cat# CBP60439 |
| HCT116 | COBIOER | Cat# CBP60028 |
| HEK293T GAS41 KO | This paper | N/A |
| HEK293T MYC KO | This paper | N/A |
| HEK293T HDAC1 KO | This paper | N/A |
| HEK293T SIN3A KO | This paper | N/A |
| HCT116 GAS41 KO | This paper | N/A |
| HCT116 GAS41 KO stably expressing GFP-Flag-GAS41 | This paper | N/A |
| HCT116 GAS41 KO stably expressing GFP-Flag-GAS41 Y74A&W93A | This paper | N/A |
| HCT116 GAS41 KO stably expressing GFP-Flag vector | This paper | N/A |
| HCT116 GAS41 KO stably expressing 3xFlag-GAS41 | This paper | N/A |
| HCT116 GAS41 KO stably expressing 3xFlag-GAS41 H43S | This paper | N/A |
| HCT116 GAS41 KO stably expressing 3xFlag-vector | This paper | N/A |
| Experimental Models: Organisms/Strains | ||
| Mouse | Taconic, Germantown, NY | NOD-SCID, female, 6-8 weeks old. |
| Oligonulceotides | ||
| Oligonucleotides used in this study | This paper | Table S4 |
| Recombinant DNA | ||
| Modified pET32a_GAS41 (a.a. 11-150) | This paper | N/A |
| Modified pET32a_GAS41 (a.a. 11-150) W93A | This paper | N/A |
| Modified pET32a_GAS41 (a.a. 11-150) Y74A/W93A | This paper | N/A |
| Modified pET32a_GAS41 (a.a. 11-150) Y139A | This paper | N/A |
| Modified pET32a_GAS41 (a.a. 11-150) del 123-128 | This paper | N/A |
| Modified pET32a_GAS41 (a.a. 11-165) | This paper | N/A |
| Modified_pET32a_MYC (a.a. 1-262) | This paper | N/A |
| pET21a_GAS41 (a.a. 11-150) | This paper | N/A |
| Modified pET15b_hH2A type 1-B/E | This paper | N/A |
| Modified pET15b_hH2B type 1-J | This paper | N/A |
| Modified pET15b_hH3.1 | This paper | N/A |
| pCR2.1 TOPO_hH3.1_K27amber | This paper | N/A |
| Modified pET15b_hH4 | This paper | N/A |
| Modified pET24_KacRS_6mt | Wakamori et al. (2015)61 | N/A |
| Modified pET15b_Kcr-RS | This paper | N/A |
| Modified pUC_one-half of a 147-bp palindromic human α-satellite DNA | Davey et al. (2002)62 | N/A |
| pRK793: TEV protease | Kapust et al. (2001)63 | Addgene Plasmid #8827 |
| CbF_Flag-GAS41 | Robert G. Roeder | N/A |
| CbF-vector | This paper | N/A |
| CbF_Flag-GAS41 N (a.a.1-165) | This paper | N/A |
| CbF_Flag-GAS41 C (a.a.148-227) | This paper | N/A |
| CbF_Flag-GAS41 del 71-75aa | This paper | N/A |
| CbF_Flag-GAS41 Y74A | This paper | N/A |
| CbF_Flag-GAS41 W93A | This paper | N/A |
| CbF_Flag-GAS41 Y74A&W93A | This paper | N/A |
| CbF_Flag-GAS41 H43A | This paper | N/A |
| CbF_Flag-GAS41 H43S | This paper | N/A |
| CbF_Flag-GAS41 Y139A | This paper | N/A |
| CbF-myc-SIN3A | This paper | N/A |
| pEGFP-C1 | Clonetech | Cat#6084-1 |
| pEGFP-MYC | This paper | N/A |
| pEGFP-MYC MBI | This paper | N/A |
| pEGFP-MYC MBII | This paper | N/A |
| pEGFP-MYC MBIII | This paper | N/A |
| pEGFP-MYC MBII+III | This paper | N/A |
| pEGFP-MYC MBIV+BZ | This paper | N/A |
| pEGFP-GAS41 | This paper | N/A |
| pCAG-GFP-Flag vector | This paper | N/A |
| pCAG-GFP-Flag-GAS41 | This paper | N/A |
| pCAG-GFP-Flag-GAS41 Y74A&W93A | This paper | N/A |
| pCAG-GFP-HDAC1 | This paper | N/A |
| pCDNA3.1(−) | ThermoFisher Scientific | Cat#V79520 |
| pcDNA3.1-myc-MYC | This paper | N/A |
| pcDNA3.1-Flag-MYC | This paper | N/A |
| pcDNA3.1-Flag-HDAC1 | This paper | N/A |
| pCAG-3xFlag vector | This paper | N/A |
| pCAG-3xFlag-GAS41 | This paper | N/A |
| pCAG-3xFlag-GAS41 H43S | This paper | N/A |
| pCAG-3xFlag-GAS41 Y139A | This paper | N/A |
| pSpCas9(BB)-2A-GFP | Addgene: PX458 | Addgene |
| Software and Algorithms | ||
| FlowJo | FLOWJO, LLC | https://www.flowjo.com |
| Image J | NIH | https://imagej.nih.gov/ij/ |
| ModFit LT | ModFit LT | https://modfit-lt.software.informer.com/ |
| Prism 8 | GraphPad | https://www.graphpad.com |
| Origin 7.0 | OriginLab | https://www.originlab.com |
| Igor Pro 8 | WaveMetrics | https://www.wavemetrics.com/ |
| Modeller (v. 9.16) | Webb & Sali (2016)64 | https://salilab.org/ |
| iMOSFLM | Battye et al. (2011)65 | https://www.mrc-lmb.cam.ac.uk/mosflm/imosflm/ver740/introduction.html |
| BALBES | Long et al. (2008)66 | https://www2.mrc-lmb.cam.ac.uk/groups/murshudov/content/balbes/balbes_layout.html |
| Phenix.refinement | Afonine et al. (2012)67 | https://phenix-online.org/ |
| COOT | Emsley et al. (2010)68 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| PyMOL | Schrödinger, LLC | https://pymol.org/2/ |
| NMRPipe | Delaglio et al. (1995)69 | https://www.ibbr.umd.edu/nmrpipe/install.html |
| NMRViewJ | Johnson & Blevins (1994)70 | https://nmrfx.org/nmrfx/nmrviewj |
| HDExaminer | Sierra Analytics | https://www.leaptec.com/ |
| MassLynx 4.2 | Waters | https://www.waters.com/nextgen/us/en.html |
| Trimmomatic v0.36 | Bolger et al. (2014)71 | http://www.usadellab.org/cms/?page=trimmomatic |
| Bowtie v1.0.1 | Langmead et al. (2009)72 | http://bowtie-bio.sourceforge.net/index.shtml |
| IGV tools v2.3.32 | Robinson et al. (2011)73 | http://www.broadinstitute.org/igv/ |
| MACS v2.1.1 | Zhang et al. (2008)74 | https://macs3-project.github.io/MACS/ |
| Bedtools v2.21.0 | Quinlan et al. (2010)75 | https://bedtools.readthedocs.io/en/latest/index.html |
| DESeq2 v1.16.1 | Love et al. (2014)76 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| TopHat v2.1.0 | Kim et al. (2013)77 | https://anaconda.org/bioconda/tophat |
| HT-seq v0.6.0 | Anders et al. (2014)78 | https://htseq.readthedocs.io/en/master/index.html |
| Other | ||
| HisTrap™ FF column, 5ml | GE Healthcare | Cat#17-0921-02 |
| HisTrap™ HP column, 5ml | GE Healthcare | Cat#17-5247-01 |
| Superdex 75 26/60 | GE Healthcare | Cat#28-9893-34 |
| Superdex 200 16/60 | GE Healthcare | Cat#28-9893-35 |
| Superdex 200 Increase 10/300L GL | GE Healthcare | Cat# 28-9909-44 |
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Cell culture
HEK293T cells and wild type HCT116 (p53+/+) cells were cultured in DMEM (Gibco) and RPMI-1640 (Gibco) contained 10% fetal calf serum (Gibco), 100 units of penicillin (Gibco) and 100 μg/ml streptomycin (Gibco).
METHOD DETAILS
Constructs
GFP-GAS41, GFP-MYC and its segment constructs of MbI (residues 1–77), MbII (residues 73–146), MbIII (residues 145–289), MbII+III (residues 73–289) and MbIV-BZ (residues 298–439) were prepared by inserting the appropriate coding DNA fragments into XhoI/BamHI sites of pEGFP-C1 vector. Flag-GAS41 in CbF vector was kindly provided by Robert Roeder (Rockefeller University). GAS41 N (residues1–165) and GAS41 C (residues148–227) were sub-cloned into EcoRV/ClaI sites of CbF vector to generate Flag-GAS41 N and Flag-GAS41 C constructs. DNA coding fragments for Flag-GAS41 and HDAC1 were inserted into the pCAG-GFP-IB vector83 to generate GFP-Flag-GAS41 and GFP-HDAC1 constructs. DNA coding fragments for GAS41 and SIN3A were inserted into modified pCAG-3xFlag vector and Cbf vector to generate 3xFlag-GAS41 and myc-SIN3A constructs. MYC was sub-cloned into XhoI/BamHI sites of lab modified pCDNA 3.1(−) vector with Flag or myc tag. The cDNAs encoding human GAS41 YEATS domain (residues 11–150, 11–165) and MYC (residues 1–262, 73–289) were cloned into the modified pET32a vectors. All mutant constructs were generated by QuickChange Site-Directed Mutagenesis Kit (Agilent Technologies). All constructs were confirmed by DNA sequencing.
Antibodies
Antibodies against H3K27ac (ab4729), RbAp48 (ab79416), MYC (ab32072), NCoR (ab24552), MBD2 (ab188474), MBD3 (ab157464), CHD3 (ab109195), p21 (ab109199), RBP1 (ab154881) and GAPDH (ab9485) were purchased from Abcam. Anti-H3K27cr (PTM-Bio 526) antibody was from PTM Biolabs Inc. Antibodies against GFP (AT0028 and AT0044), Flag-tag (AT0022 and AT0502) and myc-tag (AT0045 and AT0023) were from CMCTAG. Antibody against 6xHis-tag(D191001) was from BBI Life Sciences. Antibody against Flag-tag (F1804) was from Sigma. Anti-SIN3A (8056), anti-H3K27me3 (9733), anti-H2A.Z (50722), and normal rabbit IgG (2729) were from Cell Signaling Technologies. Anti-HDAC1 (40967) was from Active motif. Anti-HDAC2 (16152–1-AP), and anti-alpha-tubulin (66031–1-Ig) and anti-TIP60 (10827–1-AP) were obtained from ProteinTech. Anti-GAS41 (sc-393708), anti-MYC (sc-40) and normal mouse IgG (sc-2025) were from Santa Cruz. Antibody against GAS41 (GTX16452) was from Genetex. Anti-β-actin antibody (AA128) was from Beyotime. Anti-Rabbit (ab97080) and anti-mouse (ab97040) antibodies conjugated to horseradish peroxidase were from Abcam.
Generation of stable cell lines
Transfection of siRNAs was performed according to manufacturer’s instructions using RNAimax (Invitrogen) respectively. Non-targeting control, GAS41 and MYC siRNAs were purchased from GenePharma and oligonucleotides sequences are listed in Table S4. Transfection of the plasmids was performed according to manufacturer’s instructions using polyethylenimine pH7.0 (Polysciences). For generation of stable cell lines, HCT116 GAS41 KO cells were transfected with GFP-Flag-tagged and 3xFlag tagged constructs. After 72 hours, cells were cultured in selection medium containing 10 μg/ml blasticidin (Selleck) for two weeks. Expression levels of GFP fusions were determined by fluorescence microscopy and western blotting. Crotonyl-CoA was purchased from Sigma (Cat. 28007, lot BCBZ4793). Doxorubicin (S1208) and Idarubicin (S1228) and Nutlin-3a (S8059) were purchased from Selleck Chemicals.
Co-immunoprecipitation
Co-immunoprecipitation (Co-IP) using Dynabeads® Protein G (Thermo Fisher Scientific) or GFP-Trap (ChromoTek) was performed according to manufacturer’s instructions with modifications. Briefly, cells were collected, washed with cold PBS and then lysed with lysis buffer (10 mM Tris/Cl pH 7.5 containing 150 mM NaCl, 2mM MgCl2, 0.5% NP-40) containing Bezonase (Sigma), 2 mM PMSF and 1×protease inhibitor (Transgen)) at RT for 10 minutes and 4°C for 20 minutes. Supernatant was diluted with equal volume of IP binding buffer (10 mM Tris/Cl pH 7.5 containing 150 mM NaCl, 0.5 mM EDTA) and incubated with primary antibodies overnight at 4°C with constant mixing and then incubated with Dynabeads® Protein G beads slurry for additional 2 hours. For Co-IP with GFP-Trap, supernatant was incubated with GFP-Trap beads slurry for 2 hours at 4°C with rotating. Beads were then washed three times with IP wash buffer (10 mM Tris/Cl pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 0.02% Tween-20) and the bound proteins were eluted with Laemmli sample buffer and analyzed by western blot using SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific) or Veriblot for IP detection reagent (ab131366) from Abcam. For Co-IP assay assessing GAS41 mutant binding to H3K27ac or H3K27cr, cells were seeded on p100 plate to be 50% confluent at transfection. Plasmid DNA was transfected into cells with polyethylenimine. After 24 hours, cells were harvested and lysed with RIPA buffer (150mM NaCl, 50mM Tris/Cl pH 8.0, 5mM EDTA, 1% NP-40, 0.5% deoxycholate, 0.1% SDS) containing Bezonase (Sigma), 2 mM PMSF and 1×protease inhibitor (Transgen) at 4°C for 15 minutes and RT for 30 minutes. After centrifugation, supernatant was diluted with equal volume of IP binding buffer (20 mM Tris/Cl pH 8.0, 150 mM NaCl, 0.5 mM EDTA) and incubated with anti-Flag antibody overnight at 4°C with constant mixing followed by incubating with Dynabeads® Protein G beads slurry for additional 2 hours. Beads were then washed twice with IP wash butter (20 mM Tris/Cl pH 8.0, 150 mM NaCl, 0.5 mM EDTA, 0.01% Tween-20), and the bound proteins were eluted with Laemmli sample buffer followed by western blot analysis using corresponding antibodies.
Nucleosome binding experiment
HEK293T cells were seeded on p100 plate to be 50% confluent at transfection. Plasmid DNA was transfected into cells with polyethylenimine. After 24 hours, cells were harvested and lysed with IP lysis buffer (10 mM Tris/Cl pH 7.5, 150 mM NaCl, 2 mM MgCl2, 0.5% NP-40) containing Bezonase (Sigma), 2 mM PMSF and 1×protease inhibitor (Transgen) at RT for 10 minutes and 4°C for 20 minutes. After centrifugation, supernatant was diluted with IP binding buffer (10 mM Tris/Cl pH 7.5, 150 mM NaCl, 0.5 mM EDTA) containing H3K27, H3K27ac or H3K27cr peptide at indicated concentrations. Then incubated with IgG or anti-Flag antibody overnight at 4°C with constant mixing followed by incubating with Dynabeads® Protein G beads slurry for additional 2 hours. Beads were then washed twice with high salt IP wash buffer (10 mM Tris/Cl pH 7.5, 300 mM NaCl, 0.5 mM EDTA, 0.02% Tween-20) and once with low salt IP wash butter (10 mM Tris/Cl pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 0.02% Tween-20), the bound proteins were eluted with Laemmli sample buffer followed by western blot analysis using SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific).
Generation of knockout cell lines by CRISPR/Cas9
GAS41, MYC, SIN3A and HDAC1 knockout cell lines and GAS41 Y74A mutant cell line were generated using CRISPR/Cas9 method as described.84 Briefly, 20 nt oligonucleotides for Cas9 targeting were designed using CRISPR Design Tool (http://tools.genome-engineering.org) and inserted into pSpCas9(BB)-2A-GFP (Addgene, PX458) to construct Cas9 targeting plasmids. To generate GAS41, MYC, SIN3A, and HDAC1 knockout cell lines, Cas9 targeting plasmid DNA was transfected into HEK293T or HCT116 p53+/+ cells for generating the corresponding knockout cell lines. To generate GAS41 Y74A mutant cell line, Cas9 targeting plasmid DNA was co-transfected into HEK293T cells with 149 nt single-stranded DNA oligonucleotides (ssODNS) that contains flanking sequences of 73 bp on each side homologous to target region. 72 hours after transfection, individual GFP positive cells were sorted into 96 well plates using BD Influx™ cell sorter and different colonies were expanded for genomic DNA isolation. Mutations were confirmed by PCR, enzyme digestion and DNA sequencing. To sequence GAS41 alleles of Y74A heterogeneous mutant, amplicons over the Y74A targeting region were obtained by PCR and ligated into pEasy-blunt zero vector (Transgen). Thirty six colonies were sequenced to examine the heterogeneity of Y74A alleles, of which 8 are Y74A mutant and the rest are GAS41 KO. The ratio of Y74A allele to KO allele is 1:3.5. Sequences of oligonucleotides and ssODNs were listed in Table S4.
cDNA microarray and quantitative real-time PCR (qPCR)
Total RNA was isolated using TRIzol reagent (Invitrogen) according to manufacturer’s instruction followed by reverse transcription with EasyScript First-Strand cDNA Synthesis SuperMix kit (Transgen). All qPCR was performed using power SYBR green PCR master mix (Applied Biosystems) on LightCycler480 (Roche) qPCR system. The qPCR results were analyzed according to the comparative threshold cycle (Ct) method, normalized to an endogenous glyceraldehyde phosphate dehydrogenase (GAPDH) as reference and relative to an experimental control, the fold of relative expression level is given by 2−ΔΔCt85. The primer sequences for qPCR were listed in Table S4. Human colorectal cancer cDNA microarray was purchased from Shanghai Outdo Biotech Co., Ltd. (Shanghai, China), which contains cDNA from paired cancer and adjacent non-cancerous tissues. Samples preparation was briefly described. Non-cancer and cancer tissues from patients were frozen in liquid nitrogen, and tissues were cut into 3mm x 3mm x 3mm pieces on dry ice and transferred into Trizol reagent. After homogenization, total RNA was isolated according to manufacturer’s instructions and reverse-transcribed into cDNA.
ELISA assay
GAS41 YEATS domain (residues 11–150) was subcloned into the modified pET vector containing N-terminal thioredoxin-His-Tag and individual mutants were prepared using site-directed mutagenesis method. Thioredoxin-His-tag GAS41 YEATS domain and various mutants were purified with HiTrap IMAC FF column followed by Superdex 75 column (GE Healthcare). Biotin-tagged H3K27, H3K27ac, and H3K27cr peptides (residues 21–33) were loaded into Streptavidin-coated 96-well plate (Thermo Fisher Scientific) and the plate was further blocked with 1% BSA in PBS pH7.4 buffer. After washing the individual wells with PBS pH 7.4 buffer, thioredoxin-His-Tag YEATS domain or its mutants were applied into individual wells and their binding to H3K27 peptides were examined by primary His-Tag antibody followed by horseradish peroxidase conjugated secondary antibody using 3,3’,5,5’-tetramethyl-benzidine (TMB) as the substrate.
Size exclusion chromatography
HCT116 cells were lysed by RIPA buffer (150 mM NaCl, 50 mM Tris/Cl pH 8.0, 5 mM EDTA, 1% NP-40, 0.5% deoxyclolate, 0.1% SDS) and the supernatant separated from cell lysate was applied onto Superdex 200 Increase 10/300L GL column for size exclusion chromatography study. The individual fractions eluted from the column with PBS pH 6.5 buffer containing 2.0 mM EDTA, 2.0 mM DTT were resolved by SDS-PAGE and further analyzed by immunoblot using antibodies against GAS41, MYC, SIN3A and HDAC1.
Chromatin immunoprecipitation (ChIP)
Cells cultured in plates were chemically crosslinked with 1% Formaldehyde in PBS buffer for 10 minutes at room temperature followed by the addition of 2.5 M glycine (a final concentration of 125 mM) for 5 minutes. Cells (3×107) were harvested and resuspended in 1.0 ml ChIP lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris/Cl pH 8.0, protease inhibitors (Roche) and 2.0 mM PMSF). Cell suspension was sonicated for 5×60 seconds with 1-second on and 5-second off interval, at a setting of 30% duty. Sonicated chromatin was cleared by centrifuging at 20,000 g for 15 minutes at 4 °C. The cleared cell lysate was diluted 10-fold in ChIP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris/Cl pH 8.0, 167 mM NaCl) and incubated with appropriate primary antibody overnight, followed by incubated with Dynabeads® Protein G for 4 hours at 4°C. Dynabeads were washed one time with low-salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris/Cl, pH 8.0, 150 mM NaCl), high-salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM, Tris/Cl, pH 8.0, 500 mM NaCl), LiCl wash buffer (0.25 MLiCl, 1% IGEPAL-CA630, 1% deoxycholic acid (sodium salt), 1 mM EDTA, 10 mM Tris-HCl, pH 8.0) and TE buffer (10 mM Tris/Cl pH 8.0, 1 mM EDTA) for 5 minutes on a rotation platform. The immuno-precipitated complex was eluted with elution Buffer (1% SDS, 0.1 M NaHCO3) twice for 15 minutes at 65°C and the reverse cross-linking was performed at 65°C overnight. The eluted fraction was treated with RNaseA (EN0531, Thermo) for 30 min at 37°C and proteinase K (39450–01-6, Roche) for 2 hours at 65°C and extracted by phenol/chloroform and ethanol precipitated. Purified DNA was analyzed by qPCR as described above. Percent input method was used to normalize qPCR data.
ChIP-seq and ChIP-seq analysis
ChIP-sequencing were performed either on the Illumina HiSeq 2500v4 platform or BGISEQ-500 platform. The raw reads from both input and ChIP samples were filtered and trimmed using Trimmomatic,71 and then aligned to hg19 human reference genome using Bowtie72 with the –m 1 option. The peaks in the ChIP sample in reference to the input sample were called from read alignments by the MACS algorithm (v2.1.1).86 Genes associated with peaks were annotated using Homer. Peaks were chosen with the criteria FDR < 0.01 and p-value < 1×10−5. To create heatmaps, reads within regions were counted using Bedtools.75 Differential occupancy was calculated from these read counts using DESeq2.87 TSS plots were created using a combination of in-house scripting as well as ngsplot. Finally, the alignment and coverage of ChIP-seq data were visualized by integrative genomics viewer (IGV)73 (http://www.broadinstitute.org/igv/). KEGG enrichment analysis of the identified genes was performed using OmicStudio tools at https://www.omicstudio.cn/tool.
RNA-seq and RNA-seq analysis
RNA samples were prepared as described above. RNA-sequencing was performed using a BGISEQ-500 platform. Three biological replicates were prepared for each treatment. Raw reads were filtered and trimmed using Trimmomatic and then aligned to the hg19 human reference genome and indexes based on UCSC annotations using tophat. HT-Seq was used to find the read counts across the UCSC reference genome.88 Differentially expressed genes were identified by the R package DESeq2 using a false discovery rate (FDR) < 0.1 and fold-change >1.5.87 Heat maps were derived by a z-transformation of the variance-stabilized counts and sorting all genes with counts >=5 by log-fold change for each condition or treatment.
Re-ChIP
Re-ChIP was performed according to manufacturer’s instruction (53016, Active Motif). Briefly, first ChIP reaction is to incubate anti-Flag antibody with sheared chromatin prepared from formaldehyde-fixed HCT116 cells ectopically expressing Flag-GAS41. Protein-chromatin complexes are precipitated by Dynabeads® Protein G, then ‘First ChIP’ chromatin was eluted with a specialized buffer that prevents the first antibody from participating in the second ChIP reaction. The second ChIP reactions are carried out using the ‘First ChIP’ chromatin with anti-H3K27ac, anti-H3K27cr, anti-MYC, anti-SIN3A and anti-HDAC1 antibodies, respectively. After precipitation with magnetic Protein G beads, washing and reverse crosslinking, bound DNA is released, purified and analyzed by PCR to examine the co-localization for the proteins of interest at p21 gene locus.
Flow cytometry analysis
Cells were suspended as single cell dispersions and fixed with 70% ethanol at −20°C overnight, then were washed twice with PBS buffer and re-suspended in propidium iodide solution (Beijing Dingguo) containing 0.3 mg/ml RNase A (Sigma) followed by incubation at 37°C for 1 hour. Data was collected by BD FACS Calibur Cytometer and further analyzed using ModFit LT and FlowJo software programs.
Protein and peptide preparation
GAS41 YEATS domain (residues 11–150) and its mutants were expressed in E. coli BL21 (DE3) codon plus RIL strain cells as His-Tag fusion protein. After Ni column chromatography followed by thrombin cleavage for tag removal, GAS41 proteins were further purified via size-exclusion chromatography in 20 mM MES buffer of pH 6.5, containing 500 mM ammonium acetate. Uniformly 15N- and 15N/13C-labeled proteins were prepared from bacterial cells grown in M9 minimal medium containing 15NH4Cl with or without 13C6-glucose in H2O. MYC (residues 73–289) was expressed in E. coli BL21 (DE3) cells as His-MBP-Tag fusion protein. The protein was purified using Ni column chromatography and size-exclusion chromatography in 30 mM HEPES buffer of pH 7.4, containing 400 mM NaCl. After tag removal by HRV3C cleavage, MYC protein was further purified by size-exclusion chromatography in 20 mM Tris buffer of pH 8.0, containing 150 mM NaCl and 2 M urea, and followed by anion-exchange chromatography. The synthetic histone peptides were synthesized by Mimotopes company and confirmed by LC-MS analysis.
Reconstitution of H3K27-crotonylated or acetylated nucleosome core particles (NCPs)
The recombinant human histone H3 containing K27ac or K27cr was synthesized by genetic code reprogramming, as previously described61. Briefly, the cDNA encoding human histone H3.1, with the codon for K27 replaced with the TAG triplet and a terminal TAA stop codon, was subcloned into a pCR2.1-TOPO plasmid (ThermoFisher Scientific, K450002). The H3.1 protein was designed to have a histidine-rich affinity tag at the N-terminus, followed by a TEV protease recognition sequence and a linker sequence (MKDHLIHNHHKHEHAHAHEHLYFQGSSGSSG). When cleaved by TEV protease, the N-termini of K27-acetylated or K27-crotonylated H3 had a GSSGSSG linker sequence. This template plasmid (16–64 μg/mL) was used in a coupled transcription–translation cell-free system with reaction solution (9 mL) dialyzed against external feeding solution (90 mL). The reaction solution (9 mL) contained 0.37 volume of the low-molecular weight creatine phosphate tyrosine (LMCPY) mixture (160 mM HEPES-KOH buffer of pH 7.5 containing 4.1 mM L-tyrosine, 3.5 mM ATP, 2.4 mM each of GTP, CTP, UTP, 0.22 mM folic acid, 1.8 mM cAMP, 74 mM ammonium acetate, 210 mM creatine phosphate, 5 mM DTT, 530 mM potassium L-glutamate, and 11% PEG8000), 0.075 volume of the 19-amino acid mixture (20 mM each of the amino acids other than L-tyrosine), 0.01 volume of 17.5 mg/mL tRNA, 0.26 volume of E. coli S30 extract from RFzero strains60, 14 mM Mg(OAc)2, 20 mM acetyllysine, 10 mM nicotinamide, 0.05% NaN3, 250 μg/mL creatine kinase, 10 μM pyrrolysine-specific tRNA (tRNAPyl)61, 2–4 μM pyrrolysyl-tRNA synthetase (PylRS) mutant KacRS_6mt61, 67 μg/mL T7 RNA polymerase, and the template plasmid. The external feeding solution (90 mL) contained 0.03 volume of 10× S30 buffer (100 mM Tris acetate buffer of pH 8.2 containing 600 mM potassium acetate, 160 mM Mg(OAc)2, and 10 mM DTT), 0.37 volume of the LMCPY mixture, 0.075 volume of the 19-amino acid mixture, 14 mM Mg(OAc)2, 20 mM acetyllysine (or crotonyllysine), 10 mM nicotinamide, and 0.05% NaN3. Cell-free synthesis in the dialysis mode was performed at 37°C for 18 hrs, using 30 cm2 of dialysis membrane per ml reaction solution. The reaction mixture was centrifuged for 30 mins at 30,000 g at 4°C. Protein precipitates were used for histone octamer preparation. For K27-crotonylated H3, Kcr-RS containing mutations of L274A, C313F, and Y349F in Mb-PylRS89 was used instead of KacRS_6mt.
For the preparation of unmodified histones, the cDNA containing human histone H2A type 1-B/E, H2B type 1-J, H3.1, and H4 (hereafter referred to as H2A, H2B, H3 and H4, respectively) were amplified by PCR and subcloned into a pET15b-derived plasmid in which the thrombin cleavage site of pET15b (Merck Millipore, 69661) was replaced by the TEV protease recognition site. Each histone protein was designed to have an N-terminal 6×His tag followed by the TEV protease recognition site (MGSSHHHHHHSSGEHLYFQ). When cleaved by TEV protease, the N-terminus of H3 was designed to have a GSSGSSG linker sequence. The N-termini of H2A and H4 had no linker sequence and the N-terminus of H2B had one residue of G. H2A, H2, and H3 were expressed in LB broth of E. coli BL21 (DE3) cells at 37 °C. H4 was expressed in LB broth of E. coli JM109 (DE3) cells at 37°C. When the OD600 reached 0.8, isopropyl-β-D-thiogalactopyranoside was added to a final concentration of 100 μM. The cultures were incubated for an additional 16–20 hrs and cells were collected by 3-min centrifugation at 12,000 g at 20°C. Cell pellets were resuspended in 50 mM Tris-HCl buffer (pH 8.0) containing 1 M NaCl, 1% Triton X-100, and cOmplete (EDTA-free) Protease Inhibitor Cocktail (Roche). The cells were sonicated and centrifuged for 20 mins at 18,000 g at 20°C.
The precipitated histones were solubilized by sonication in 20 mM Tris-HCl buffer (pH 8.0) containing 6 M guanidine-hydrochloride, 500 mM NaCl, and 5 mM imidazole. The sample was centrifuged for 20 mins at 18,000 g at 20°C and the supernatant was loaded onto a HisTrap HP 5ml column (GE Healthcare) equilibrated in 10 mM Tris-HCl buffer (pH 8.0) containing 500 mM NaCl, 5 mM imidazole, and 6 M urea. Histones were eluted with 10 mM Tris-HCl buffer (pH 8.0) containing 500 mM NaCl, 500 mM imidazole, and 6 M urea. The eluted histones were dialyzed against cold distilled water at 4°C overnight. The N-terminal histidine-rich tag was cleaved by TEV protease63 in 10 mM Tris-HCl buffer (pH 8.0) at a weight ratio of histone:TEV protease = 2–10:1 at 4°C overnight. The solution of cleaved histones was adjusted to contain 10mM Tris-HCl buffer (pH 8.0) containing 500 mM NaCl, 5 mM imidazole, and 6 M urea. The sample solution was then reapplied to a HisTrap HP 5ml column (GE Healthcare) and the flow-through fraction was collected. The collected protein was dialyzed against cold distilled water at 4°C overnight. Purified histones were lyophilized. The purified histones (i.e., H2A, H2B, H3, and H4) were mixed at an equimolar ratio in 20 mM Tris-HCl buffer (pH 8.0) containing 6 M guanidine-hydrochloride and 10 mM DTT and dialyzed at 4°C overnight against 10 mM Tris-HCl buffer (pH 7.6) containing 2 M NaCl, 1 mM EDTA, and 5 mM β-mercaptoethanol. Histone octamers were purified by gel filtration chromatography using a HiLoad Superdex 200 16/60 column (GE Healthcare) equilibrated in 10 mM Tris-HCl buffer (pH 7.6) containing 2 M NaCl, 1 mM EDTA, and 5 mM β-mercaptoethanol, and concentrated in Amicon Ultra-15 centrifugal filter units (Merck Millipore, 30 kDa MWCO).
The nucleosome consisted of the histone octamer and the palindromic 147-bp human α-satellite DNA was reconstituted as reported61,62. The histones in the nucleosome containing the unmodified, K27-crotonylated, or K27-acetylated H3 were analyzed by immunoblotting, using the following antibodies and dilution rates: H3 (Abcam, ab1791, 1/2,000), H3K27cr (ThermoFisher Scientific, 712478, 1/500), and H3K27ac (Millipore, 07–360, 1/2,000). Histone octamers and the 147-bp DNA were mixed at a 1:1.1 molar ratio. The mixed solution was dialyzed against 10 mM Tris-HCl buffer (pH 7.6) containing 2 M KCl, 1 mM EDTA, and 1 mM DTT for 2 hrs at 4°C. The concentration of KCl was then gradually decreased by diluting for 16 hrs with 10 mM Tris-HCl buffer (pH 7.6) containing 250 mM KCl, 1 mM EDTA, and 1 mM DTT using a peristaltic pump (ATTO, SJ-1211II-H). The solution was dialyzed against 10 mM Tris-HCl buffer (pH 7.6) containing 250 mM KCl, 1 mM EDTA, and 1 mM DTT for 2 hrs at °C. Reconstituted nucleosomes were centrifuged for 20 mins at 18,000 g at 4°C and the supernatants were collected, incubated at 55°C for 2 hrs, and stored at 4°C. Nucleosomes were concentrated in Amicon Ultra-15 centrifugal filter units (Merck Millipore, 30 kDa MWCO) and purified on a 6% native polyacrylamide gel (acrylamide:N,N’-methylenbisacrylamide = 59:1), using a Model 491 Prep Cell apparatus (Bio-Rad, 1702928) eluted with 20 mM Tris-HCl buffer (pH 7.6) containing 1 mM EDTA and 1 mM DTT. The eluted fractions were concentrated to approximately 25 μM. For microscale thermophoresis assay, H3K27cr NCP, H3K27ac NCP, and unmodified NCP were dialyzed against 20 mM Tris-HCl buffer (pH 7.6) at 4°C.
Protein crystallization and structure determination
Purified GAS41 YEATS domain (residues 11–150) was mixed with a H3K27cr peptide (residues 21–33) at 1:10 molar ratio. Crystallization was performed under 15°C via sitting-drop method under the condition of 0.1 M HEPES (pH 7.5) and 1.8 M ammonium sulfate. Diffraction data were collected at Photon Factory BL-17A (Tsukuba, Japan) under cryo-conditions. The data were processed with iMOSFLM65 and other programs from the CCP4 suite.90 Phase was solved by molecular replacement using BALBES.66 Phenix refinement67 was used for electron density map refinement. Graphic program COOT68 was used for model building and visualization. Structure figures were generated using PyMOL Molecular Graphics System (Schrödinger, LLC). Data collection and statistics of structure refinement are summarized in Table S2.
NMR spectroscopy
NMR samples of the GAS41 YEATS domain (residues 11–150) in complex with H3K27cr peptide were prepared in 20 mM MES buffer (pH 6.5) containing 200 mM ammonium acetate and 500 mM urea in H2O/2H2O (9:1) or 2H2O. All NMR spectra were collected at 20°C on NMR spectrometers of 900, 800, 600, or 500 MHz with a cryoprobe. 1H, 13C, and 15N resonances of the protein in the GAS41/H3K27cr complex were assigned with triple-resonance NMR spectra collected with the 13C/15N-labeled and 75% deuterated protein (0.5 mM) bound to the unlabeled peptide (2.5 mM).45 GAS41 YEATS domain binding to the H3K27cr peptide and purified MYC protein was carried out in 2D 1H-15N HSQC spectra of 15N-labeled GAS41 YEATS domain.
Isothermal titration calorimetry
Experiments were carried out on a MicroCal ITC200 instrument at 20°C while stirring at 750 rpm in PBS buffer (pH 7.4) buffer containing, 2.0 mM EDTA, 2.0 mM β-mercaptoethanol, and 500 mM NaCl. Peptide concentration was determined by weight and confirmed by NMR, and protein concentrations by A280 measurements. The protein sample (0.23 mM) was placed in the cell, whereas the micro-syringe was loaded with histone peptides (4.0 mM) in the same buffer as the protein sample. The titrations were conducted using 17 successive injections of 2.0 ml (the first at 0.4 l, and the remaining 16 at 2.0 μl) with a duration of 4 s per injection and a spacing of 180 s between injections. The collected data were processed using the Origin 7.0 software program (Origin Lab) supplied with the instrument according to the ‘one set of sites’ fitting model.
Tumorigenesis study
All procedures were approved by the Institutional Animal Care and Use Committee at Jilin University and conformed to the legal mandates and federal guidelines for the care and maintenance of laboratory animals. Animals were maintained and treated under pathogen-free conditions. Female NOD-SCID mice (6–8 weeks old; Taconic, Germantown, NY) were subcutaneously injected with 1×106 HCT116 wt or GAS41 knockout cells per mouse, respectively. Tumor growth was monitored with caliper measurements. When tumors were approximately 1.5 cubic centimeter in size, mice were euthanized and tumors excised. Tumor size and weight will be recorded.
Peptide pull-down assay
HCT116 cells were transfected with Flag-GAS41. After 24 hours, cells were harvested and lysed with RIPA buffer (150 mM NaCl, 50 mM Tris/Cl pH 8.0, 5 mM EDTA, 1% NP-40, 0.5% deoxycholate, 0.1% SDS) containing Bezonase (Sigma), 2 mM PMSF and 1×protease inhibitor (Transgen) at 4°C for 15 minutes and RT for 30 minutes. After centrifugation, supernatant was diluted with equal volume of IP binding buffer (20 mM Tris/Cl pH 8.0, 150 mM NaCl, 0.5 mM EDTA) and incubated with anti-Flag antibody and biotinylated histone peptides overnight at 4°C with constant mixing followed by incubating with streptavidin magnetic beads (Engibody IF9042) for additional 2 hours. Beads were then washed twice with IP wash butter (20 mM Tris/Cl pH 8.0, 150 mM NaCl, 0.5 mM EDTA, 0.01% Tween-20), and the bound proteins were eluted with Laemmli sample buffer followed by western blot analysis using corresponding antibodies.
Microscale thermophoresis (MST) assay
The MST measurements for the binding of GFP-GAS41 to H3K27ac or H3K27cr peptide and for the binding to GAS41 YEATS domain (residues 11–150) to H3K27cr NCP, H3K27ac NCP or unmodified NCP were performed using Monolith NT.115 (NanoTemper). GFP-GAS41 protein was used as 25 nM to provide optimal fluorescent signal in the binding reaction. H3K27 peptides were diluted with PBS, pH 7.4, 0.05% Tween® 20 for 16 times at 1:1 serial dilution beginning with 4 mM. GAS41 YEATS domain was used as 50 nM to provide optimal fluorescent signal in the binding reaction. The modified and unmodified NCPs were diluted with 20 mM Tris, pH 7.6, 150 mM NaCl for 16 times at 1:1 serial dilution beginning with 20 μM. After 10 min incubation at room temperature, samples were loaded into capillaries and measurement was carried out at 40% MST power and 40% excitation power using the MO Control software.
Hydrogen/deuterium exchange–mass spectrometry (HDX-MS) analysis
HDX-MS analysis was performed to determine the binding site of GAS41 in MYC. We examined differences in HDX rates of MYC using a 1.7 mg/ml MYC (residues 73–289) and an equimolar concentration of GAS41 (residues 11–150) in the H2O-based 20 mM MES (pH6.5), 200 mM NaCl, 500 mM Urea, 2 mM DTT. The HDX reaction was started by diluting D2O-based buffer by 10-fold at 10°C and then incubated separately at each time as shown in Table S1. The D2O-incubated samples were quenched by the addition of an equal volume of prechilled quenching buffer (8 M urea, 1 M Tris(2-carboxyethyl)phosphine hydrochloride, pH 3.0) with the HDx-3 PAL (LEAP Technologies). The quenched protein samples were subjected to online pepsin digestion (Enzymate Protein Pepsin Column, 30 × 2.1mm, Waters) and analyzed by LC-MS using UltiMate3000RSLCnano (Thermo Fisher Scientific) connected to the Q Exactive HF-X mass spectrometer (Thermo Fisher Scientific). Online pepsin digestion was performed in formic acid solution, pH 2.5 at 8°C for 3 min at a flow rate of 50 μL/min. The desalting column and the analytical columns were Acclaim PepMap300 C18 (1.0 × 15 mm; Thermo Fisher Scientific) and Hypersil Gold (1.0 × 50 mm; Thermo Fisher Scientific), respectively. The mobile phases were 0.1% formic acid solution (A buffer) and 0.1% formic acid containing 90% acetonitrile (B buffer). The deuterated peptides were eluted at a flow rate of 45 μL/min with a gradient of 10–90% of B buffer in 9 min. Mass spectrometer conditions were as follows: an electrospray voltage of 3.8 kV, positive ion mode, sheath and auxiliary nitrogen flow rate at 20 and 2 arbitrary units, ion transfer tube temperature at 275°C, auxiliary gas heater temperature at 100°C, and a mass range of m/z 200–2000. The data-dependent acquisition was performed with normalized collision energy of 27 arbitrary units. The MS and MS/MS spectra were subjected to a database search analysis using the Proteome Discoverer 2.2 (Thermo Fisher Scientific) against an in-house database containing the amino acid sequence of the MYC. The search results and MS raw files were used for the analysis of the deuteration levels of the peptide fragments using the HDExaminer software (Sierra Analytics). Data summary for each HDX-MS was shown in Table S1.
QUANTIFICATION AND STATISTICAL ANALYSES
Throughout this study results were presented as the mean ± s.e.m. from the number of replicates indicated in the corresponding figure legends. Statistical significance was calculated using a two-tailed paired t-test, with P < 0.05 considered statistically significant unless noted otherwise. Statistical significance levels are denoted as follows: P < 0.05 (∗), P < 0.01 (∗∗), P < 0.001 (∗∗∗), ns, not significant.
Supplementary Material
Nan et al. demonstrate that GAS41 through its YEAST domain recognizes histone H3 lysine 27 (H3K27) crotonylation and is recruited by proto-oncogenic MYC to SIN3A-HDAC1 co-repressors to repress gene transcription. These results suggest that H3K27 crotonylation, trimethylation, and acetylation represent distinct chromatin states for transcriptional repression, silencing, and activation, respectively.
H3 lysine 27 crotonylation is a chromatin state for gene transcriptional repression
GAS41 YEATS domain recognizes H3 lysine 27 crotonylation and proto-oncogenic MYC
GAS41 and MYC amplification likely causes p21 down-regulation in colorectal cancer
ACKNOWLEDGEMENTS
We thank the BL-17A beamline staff at Photon Factory and Dr. Seetharaman Jayaraman for assistance with X-ray data collection at APS/ALS, and Drs. Rinku Jain, Kyohei Arita, and Nicolas Babault for technical advice on structure refinement, and the New York Structural Biology Center for the use of the NMR facilitates. We are grateful to the Roeder Lab in the Rockefeller University for providing the CbF-GAS41 construct. This work was supported in part by the research funds from the First Hospital of Jilin University, the JLU Science and Technology Innovative Research Team Grant (2017TD-25), the Natural Science Foundation of China (31701097, N.L.), Natural Science Foundation of Jilin Province (20200201427JC, N.L.; 20180101235JC, Q.Z.), Platform Project for Supporting Drug Discovery and Life Science Research (Basis for Supporting Innovative Drug Discovery and Life Science Research (BINDS)) from AMED of Japan (JP22ama121033j0001, K.T.), Takeda Science Foundation (T.K.), and the National Institutes of Health (M.-M.Z.). We would like to dedicate this study in memory of Dr. C. David Allis (1951–2023) for his pioneering contributions in the field of chromatin biology and gene transcription.
Footnotes
DECLARATION OF INTERESTS
M.-M.Z. is a scientific founder, director and shareholder of Parkside Scientific Inc. The other authors do not declare any competing interests.
INCLUSION AND DIVERSITY
We support inclusive, diverse, and equitable conduct of research.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at http:…
Table S3 summarizes the effects of GAS41 knockdown on gene transcription in HCT116 cells, related to Figure 6.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES:
- 1.Tan M, Luo H, Lee S, Jin F, Yang JS, Montellier E, Buchou T, Cheng Z, Rousseaux S, Rajagopal N, et al. (2011). Identification of 67 Histone Marks and Histone Lysine Crotonylation as a New Type of Histone Modification. Cell 146, 1016–1028. 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, and Zhou MM (1999). Structure and ligand of a histone acetyltransferase bromodomain. Nature 399, 491–496. 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 3.Zaware N, and Zhou MM (2019). Bromodomain biology and drug discovery. Nat Struct Mol Biol 26, 870–879. 10.1038/s41594-019-0309-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schulze JM, Wang AY, and Kobor MS (2010). Reading chromatin: insights from yeast into YEATS domain structure and function. Epigenetics : official journal of the DNA Methylation Society 5, 573–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Y, Wen H, Xi Y, Tanaka K, Wang H, Peng D, Ren Y, Jin Q, Dent SY, Li W, et al. (2014). AF9 YEATS domain links histone acetylation to DOT1L-mediated H3K79 methylation. Cell 159, 558–571. 10.1016/j.cell.2014.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.L Y, Sabari BR, Panchenko T, Wen H, Zhao D, Guan H, Wan L, Huang H, Tang Z, Zhao Y, et al. (2016). Molecular Coupling of Histone Crotonylation and Active Transcription by AF9 YEATS Domain. Mol Cell 62, 181–193. 10.1016/j.molcel.2016.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wan L, Wen H, Li Y, Lyu J, Xi Y, Hoshii T, Joseph JK, Wang X, Loh YE, Erb MA, et al. (2017). ENL links histone acetylation to oncogenic gene expression in acute myeloid leukaemia. Nature 543, 265–269. 10.1038/nature21687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erb MA, Scott TG, Li BE, Xie H, Paulk J, Seo HS, Souza A, Roberts JM, Dastjerdi S, Buckley DL, et al. (2017). Transcription control by the ENL YEATS domain in acute leukaemia. Nature 543, 270–274. 10.1038/nature21688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perlman EJ, Gadd S, Arold ST, Radhakrishnan A, Gerhard DS, Jennings L, Huff V, Guidry Auvil JM, Davidsen TM, Dome JS, et al. (2015). MLLT1 YEATS domain mutations in clinically distinctive Favourable Histology Wilms tumours. Nature communications 6, 10013. 10.1038/ncomms10013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wan L, Chong S, Xuan F, Liang A, Cui X, Gates L, Carroll TS, Li Y, Feng L, Chen G, et al. (2020). Impaired cell fate through gain-of-function mutations in a chromatin reader. Nature 577, 121–126. 10.1038/s41586-019-1842-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mi W, Guan H, Lyu J, Zhao D, Xi Y, Jiang S, Andrews FH, Wang X, Gagea M, Wen H, et al. (2017). YEATS2 links histone acetylation to tumorigenesis of non-small cell lung cancer. Nature communications 8, 1088. 10.1038/s41467-017-01173-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gowans GJ, Bridgers JB, Zhang J, Dronamraju R, Burnetti A, King DA, Thiengmany AV, Shinsky SA, Bhanu NV, Garcia BA, et al. (2019). Recognition of Histone Crotonylation by Taf14 Links Metabolic State to Gene Expression. Mol Cell 76, 909–921 e903. 10.1016/j.molcel.2019.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park JH, and Roeder RG (2006). GAS41 is required for repression of the p53 tumor suppressor pathway during normal cellular proliferation. Mol Cell Biol 26, 4006–4016. 10.1128/mcb.02185-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischer U, Heckel D, Michel A, Janka M, Hulsebos T, and Meese E. (1997). Cloning of a novel transcription factor-like gene amplified in human glioma including astrocytoma grade I. Hum Mol Genet 6, 1817–1822. [DOI] [PubMed] [Google Scholar]
- 15.Tao K, Yang J, Hu Y, and Deng A. (2015). Knockdown of YEATS4 inhibits colorectal cancer cell proliferation and induces apoptosis. Am J Transl Res 7, 616–623. [PMC free article] [PubMed] [Google Scholar]
- 16.Ji S, Zhang Y, and Yang B. (2017). YEATS Domain Containing 4 Promotes Gastric Cancer Cell Proliferation and Mediates Tumor Progression Via Activating the Wnt/beta-catenin Signaling Pathway. Oncol Res. 10.3727/096504017X14878528144150. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Pikor LA, Lockwood WW, Thu KL, Vucic EA, Chari R, Gazdar AF, Lam S, and Lam WL (2013). YEATS4 is a novel oncogene amplified in non-small cell lung cancer that regulates the p53 pathway. Cancer research 73, 7301–7312. 10.1158/0008-5472.can-13-1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fischer U, Meltzer P, and Meese E. (1996). Twelve amplified and expressed genes localized in a single domain in glioma. Human genetics 98, 625–628. [DOI] [PubMed] [Google Scholar]
- 19.Jixiang C, Shengchun D, Jianguo Q, Zhengfa M, Xin F, Xuqing W, Jianxin Z, and Lei C. (2017). YEATS4 promotes the tumorigenesis of pancreatic cancer by activating beta-catenin/TCF signaling. Oncotarget 8, 25200–25210. 10.18632/oncotarget.15633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doyon Y, Selleck W, Lane W, Tan S, and Cote J. (2004). Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol Cell Biol 24, 1884–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cai Y, Jin J, Florens L, Swanson SK, Kusch T, Li B, Workman JL, Washburn MP, Conaway RC, and Conaway JW (2005). The mammalian YL1 protein is a shared subunit of the TRRAP/TIP60 histone acetyltransferase and SRCAP complexes. J Biol Chem 280, 13665–13670. 10.1074/jbc.M500001200. [DOI] [PubMed] [Google Scholar]
- 22.Piccinni E, Chelstowska A, Hanus J, Widlak P, Loreti S, Tata AM, Augusti-Tocco G, Bianchi MM, and Negri R. (2011). Direct interaction of Gas41 and Myc encoded by amplified genes in nervous system tumours. Acta biochimica Polonica 58, 529–534. [PubMed] [Google Scholar]
- 23.Heisel S, Habel NC, Schuetz N, Ruggieri A, and Meese E. (2010). The YEATS family member GAS41 interacts with the general transcription factor TFIIF. BMC Mol Biol 11, 53. 10.1186/1471-2199-11-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ding X, Fan C, Zhou J, Zhong Y, Liu R, Ren K, Hu X, Luo C, Xiao S, Wang Y, et al. (2006). GAS41 interacts with transcription factor AP-2beta and stimulates AP-2beta-mediated transactivation. Nucleic Acids Res 34, 2570–2578. 10.1093/nar/gkl319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Debernardi S, Bassini A, Jones L, Chaplin T, Linder B, de Bruijn D, Meese E, and Young B. (2002). The MLL fusion partner AF10 binds GAS41, a protein that interacts with the human SWI/SNF complex. Blood 99, 275–281. [DOI] [PubMed] [Google Scholar]
- 26.Lauffart B, Howell SJ, Tasch JE, Cowell JK, and Still IH (2002). Interaction of the transforming acidic coiled-coil 1 (TACC1) protein with ch-TOG and GAS41/NuBI1 suggests multiple TACC1-containing protein complexes in human cells. Biochem J 363, 195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zimmermann K, Ahrens K, Matthes S, Buerstedde JM, Stratling WH, and Phi-van L. (2002). Targeted disruption of the GAS41 gene encoding a putative transcription factor indicates that GAS41 is essential for cell viability. J Biol Chem 277, 18626–18631. 10.1074/jbc.M200572200. [DOI] [PubMed] [Google Scholar]
- 28.Park JH, Smith RJ, Shieh SY, and Roeder RG (2011). The GAS41-PP2Cbeta complex dephosphorylates p53 at serine 366 and regulates its stability. J Biol Chem 286, 10911–10917. 10.1074/jbc.C110.210211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hsu CC, Zhao D, Shi J, Peng D, Guan H, Li Y, Huang Y, Wen H, Li W, Li H, and Shi X. (2018). Gas41 links histone acetylation to H2A.Z deposition and maintenance of embryonic stem cell identity. Cell Discov 4, 28. 10.1038/s41421-018-0027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hsu CC, Shi J, Yuan C, Zhao D, Jiang S, Lyu J, Wang X, Li H, Wen H, Li W, and Shi X. (2018). Recognition of histone acetylation by the GAS41 YEATS domain promotes H2A.Z deposition in non-small cell lung cancer. Genes Dev 32, 58–69. 10.1101/gad.303784.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andrews FH., Strahl BD, and Kutateladze TG (2016). Insights into newly discovered marks and readers of epigenetic information. Nat Chem Biol 12, 662–668. 10.1038/nchembio.2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Q, Zeng L, Zhao C, Ju Y, Konuma T, and Zhou MM (2016). Structural Insights into Histone Crotonyl-Lysine Recognition by the AF9 YEATS Domain. Structure 24, 1606–1612. 10.1016/j.str.2016.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kikuchi M, Morita S, Goto M, Wakamori M, Katsura K, Hanada K, Shirouzu M, and Umehara T. (2022). Elucidation of binding preferences of YEATS domains to site-specific acetylated nucleosome core particles. J Biol Chem 298, 102164. 10.1016/j.jbc.2022.102164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.El-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, and Vogelstein B. (1993). WAF1, a potential mediator of p53 tumor suppression. Cell 75, 817–825. 10.1016/0092-8674(93)90500-P. [DOI] [PubMed] [Google Scholar]
- 35.Sabari BR, Tang Z, Huang H, Yong-Gonzalez V, Molina H, Kong HE, Dai L, Shimada M, Cross JR, Zhao Y, et al. (2015). Intracellular crotonyl-CoA stimulates transcription through p300-catalyzed histone crotonylation. Mol Cell 58, 203–215. 10.1016/j.molcel.2015.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cowley SM, Iritani BM, Mendrysa SM, Xu T, Cheng PF, Yada J, Liggitt HD, and Eisenman RN (2005). The mSin3A chromatin-modifying complex is essential for embryogenesis and T-cell development. Mol Cell Biol 25, 6990–7004. 10.1128/mcb.25.16.6990-7004.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dannenberg JH, David G, Zhong S, van der Torre J, Wong WH, and Depinho RA (2005). mSin3A corepressor regulates diverse transcriptional networks governing normal and neoplastic growth and survival. Genes Dev 19, 1581–1595. 10.1101/gad.1286905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li D-Q, Pakala SB, Reddy SDN, Ohshiro K, Peng S-H, Lian Y, Fu SW, and Kumar R. (2010). Revelation of p53-independent Function of MTA1 in DNA Damage Response via Modulation of the p21(WAF1)-Proliferating Cell Nuclear Antigen Pathway. The Journal of Biological Chemistry 285, 10044–10052. 10.1074/jbc.M109.079095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bailey P, Downes M, Lau P, Harris J, Chen SL, Hamamori Y, Sartorelli V, and Muscat GE (1999). The nuclear receptor corepressor N-CoR regulates differentiation: N-CoR directly interacts with MyoD. Molecular endocrinology (Baltimore, Md.) 13, 1155–1168. 10.1210/mend.13.7.0305. [DOI] [PubMed] [Google Scholar]
- 40.Wu S, Cetinkaya C, Munoz-Alonso MJ, von der Lehr N, Bahram F, Beuger V, Eilers M, Leon J, and Larsson LG (2003). Myc represses differentiation-induced p21CIP1 expression via Miz-1-dependent interaction with the p21 core promoter. Oncogene 22, 351–360. 10.1038/sj.onc.1206145. [DOI] [PubMed] [Google Scholar]
- 41.Herold S, Wanzel M, Beuger V, Frohme C, Beul D, Hillukkala T, Syvaoja J, Saluz HP, Haenel F, and Eilers M. (2002). Negative regulation of the mammalian UV response by Myc through association with Miz-1. Mol Cell 10, 509–521. 10.1016/s1097-2765(02)00633-0. [DOI] [PubMed] [Google Scholar]
- 42.Nair SK., and Burley SK (2003). X-ray structures of Myc-Max and Mad-Max recognizing DNA. Molecular bases of regulation by proto-oncogenic transcription factors. Cell 112, 193–205. [DOI] [PubMed] [Google Scholar]
- 43.Cho HJ, Li H, Linhares BM, Kim E, Ndoj J, Miao H, Grembecka J, and Cierpicki T. (2018). GAS41 Recognizes Diacetylated Histone H3 through a Bivalent Binding Mode. ACS Chem Biol 13, 2739–2746. 10.1021/acschembio.8b00674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Andrews FH, Shinsky SA, Shanle EK, Bridgers JB, Gest A, Tsun IK, Krajewski K, and Shi X. (2016). The Taf14 YEATS domain is a reader of histone crotonylation. Nature Chem. Bio. 12, 396–398. 10.1038/nchembio.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clore GM, and Gronenborn AM (1994). Multidimensional heteronuclear nuclear magnetic resonance of proteins. Methods in enzymology 239, 349–363. [DOI] [PubMed] [Google Scholar]
- 46.Tang Y, Luo J, Zhang W, and Gu W. (2006). Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Molecular cell 24, 827–839. 10.1016/j.molcel.2006.11.021. [DOI] [PubMed] [Google Scholar]
- 47.Bassi C, Li YT, Khu K, Mateo F, Baniasadi PS, Elia A, Mason J, Stambolic V, Pujana MA, Mak TW, and Gorrini C. (2016). The acetyltransferase Tip60 contributes to mammary tumorigenesis by modulating DNA repair. Cell Death Differ 23, 1198–1208. 10.1038/cdd.2015.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee MS, Seo J, Choi DY, Lee EW, Ko A, Ha NC, Yoon JB, Lee HW, Kim KP, and Song J. (2013). Stabilization of p21 (Cip1/WAF1) following Tip60-dependent acetylation is required for p21-mediated DNA damage response. Cell Death Differ 20, 620–629. 10.1038/cdd.2012.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Legube G, Linares LK, Tyteca S, Caron C, Scheffner M, Chevillard-Briet M, and Trouche D. (2004). Role of the histone acetyl transferase Tip60 in the p53 pathway. J Biol Chem 279, 44825–44833. 10.1074/jbc.M407478200. [DOI] [PubMed] [Google Scholar]
- 50.Yap KL, Li S, Munoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, Gil J, Walsh MJ, and Zhou MM (2010). Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell 38, 662–674. 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Strahl BD, and Allis CD (2000). The language of covalent histone modifications. Nature 403, 41–45. 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 52.Allis CD, and Jenuwein T. (2016). The molecular hallmarks of epigenetic control. Nat Rev Genet 17, 487–500. 10.1038/nrg.2016.59. [DOI] [PubMed] [Google Scholar]
- 53.Mitchell KO, and El-Deiry WS (1999). Overexpression of c-Myc inhibits p21WAF1/CIP1 expression and induces S-phase entry in 12-O-tetradecanoylphorbol-13-acetate (TPA)-sensitive human cancer cells. Cell Growth Differ 10, 223–230. [PubMed] [Google Scholar]
- 54.Claassen GF., and Hann SR (2000). A role for transcriptional repression of p21CIP1 by c-Myc in overcoming transforming growth factor beta -induced cell-cycle arrest. Proc Natl Acad Sci U S A 97, 9498–9503. 10.1073/pnas.150006697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Coller HA, Grandori C, Tamayo P, Colbert T, Lander ES, Eisenman RN, and Golub TR (2000). Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling, and adhesion. Proc Natl Acad Sci U S A 97, 3260–3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gartel AL, Ye X, Goufman E, Shianov P, Hay N, Najmabadi F, and Tyner AL (2001). Myc represses the p21(WAF1/CIP1) promoter and interacts with Sp1/Sp3. Proc Natl Acad Sci U S A 98, 4510–4515. 10.1073/pnas.081074898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Seoane J, Le HV, and Massague J. (2002). Myc suppression of the p21(Cip1) Cdk inhibitor influences the outcome of the p53 response to DNA damage. Nature 419, 729–734. 10.1038/nature01119. [DOI] [PubMed] [Google Scholar]
- 58.Gartel AL, and Shchors K. (2003). Mechanisms of c-myc-mediated transcriptional repression of growth arrest genes. Experimental cell research 283, 17–21. [DOI] [PubMed] [Google Scholar]
- 59.Wanzel M, Herold S, and Eilers M. (2003). Transcriptional repression by Myc. Trends Cell Biol 13, 146–150. 10.1016/s0962-8924(03)00003-5. [DOI] [PubMed] [Google Scholar]
- 60.Mukai T, Yanagisawa T, Ohtake K, Wakamori M, Adachi J, Hino N, Sato A, Kobayashi T, Hayashi A, Shirouzu M, et al. (2011). Genetic-code evolution for protein synthesis with non-natural amino acids. Biochem Biophys Res Commun 411, 757–761. 10.1016/j.bbrc.2011.07.020. [DOI] [PubMed] [Google Scholar]
- 61.Wakamori M, Fujii Y, Suka N, Shirouzu M, Sakamoto K, Umehara T, and Yokoyama S. (2015). Intra- and inter-nucleosomal interactions of the histone H4 tail revealed with a human nucleosome core particle with genetically-incorporated H4 tetra-acetylation. Sci Rep 5, 17204. 10.1038/srep17204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Davey CA, Sargent DF, Luger K, Maeder AW, and Richmond TJ (2002). Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 a resolution. Journal of molecular biology 319, 1097–1113. 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 63.Kapust RB, Tozser J, Fox JD, Anderson DE, Cherry S, Copeland TD, and Waugh DS (2001). Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng 14, 993–1000. 10.1093/protein/14.12.993. [DOI] [PubMed] [Google Scholar]
- 64.Webb B, and Sali A. (2016). Comparative Protein Structure Modeling Using MODELLER. Current protocols in protein science 86, 2.9.1–2.9.37. 10.1002/cpps.20. [DOI] [PubMed] [Google Scholar]
- 65.Battye TG, Kontogiannis L, Johnson O, Powell HR, and Leslie AG (2011). iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr 67, 271–281. 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Long F, Vagin AA, Young P, and Murshudov GN (2008). BALBES: a molecular-replacement pipeline. Acta Crystallogr D Biol Crystallogr 64, 125–132. 10.1107/S0907444907050172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Afonine PV., Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, and Adams PD (2012). Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr 68, 352–367. 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Emsley P, Lohkamp B, Scott WG, and Cowtan K. (2010). Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501. 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, and Bax A. (1995). NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6, 277–293. [DOI] [PubMed] [Google Scholar]
- 70.Johnson B, and Blevins R. (1994). NMRView: A computer program for the visualization and analysis of NMR data. J. Biomol. NMR 4, 603–614. [DOI] [PubMed] [Google Scholar]
- 71.Bolger AM, Lohse M, and Usadel B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Langmead B, Trapnell C, Pop M, and Salzberg SL (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10, R25. 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, and Mesirov JP (2011). Integrative genomics viewer. Nat Biotechnol 29, 24–26. 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, and Liu XS (2008). Model-based Analysis of ChIP-Seq (MACS). Genome biology 9, R137. 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Quinlan AR, and Hall IM (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Love MI, Huber W, and Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology 15, 550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, and Salzberg SL (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome biology 14, R36. 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Anders S, Pyl PT, and Huber W. (2014). HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics (Oxford, England) 31, 166–169. 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Spandidos A, Wang X, Wang H, Dragnev S, Thurber T, and Seed B. (2008). A comprehensive collection of experimentally validated primers for Polymerase Chain Reaction quantitation of murine transcript abundance. BMC Genomics 9, 633. 10.1186/1471-2164-9-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Spandidos A, Wang X, Wang H, and Seed B. (2010). PrimerBank: a resource of human and mouse PCR primer pairs for gene expression detection and quantification. Nucleic Acids Res 38, D792–799. 10.1093/nar/gkp1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang X, and Seed B. (2003). A PCR primer bank for quantitative gene expression analysis. Nucleic Acids Res 31, e154. 10.1093/nar/gng154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Donner AJ, Szostek S, Hoover JM, and Espinosa JM (2007). CDK8 is a stimulus-specific positive coregulator of p53 target genes. Mol Cell 27, 121–133. 10.1016/j.molcel.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schermelleh L, Haemmer A, Spada F, Rosing N, Meilinger D, Rothbauer U, Cardoso MC, and Leonhardt H. (2007). Dynamics of Dnmt1 interaction with the replication machinery and its role in postreplicative maintenance of DNA methylation. Nucleic Acids Res 35, 4301–4312. 10.1093/nar/gkm432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, and Zhang F. (2013). Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8, 2281–2308. 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Livak KJ, and Schmittgen TD (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods (San Diego, Calif 25, 402–408. 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 86.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, and Liu XS (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol 9, R137. 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Love MI, Huber W, and Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Anders S, Pyl PT, and Huber W. (2015). HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kim CH, Kang M, Kim HJ, Chatterjee A, and Schultz PG (2012). Site-specific incorporation of epsilon-N-crotonyllysine into histones. Angew Chem Int Ed Engl 51, 7246–7249. 10.1002/anie.201203349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, et al. (2011). Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67, 235–242. 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All structure coordinates were deposited in the Protein Data Bank (PDB) with identification number 8I60. Genomic sequencing datasets from this study were deposited to the NCBI Gene Expression Omnibus (GEO) data repository under accession number GSE128590. Source image files for figures are deposited to Mendeley Data. All data are publicly available as of the date of publication. Accession numbers and DOI are listed in the Key Resources Table.
This paper does not report any original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-H3K27ac Antibody | Abcam | Cat# ab4729, RRID:AB_2118291 |
| Rabbit anti-H3K27ac Antibody | Millipore | Cat# 07-360, RRID: AB_310550 |
| Rabbit anti-H3K27cr Antibody | ThermoFisher Scientific | Cat# 712478, RRID:AB_2848247 |
| Rabbit anti-H3 Antibody | Abcam | Cat# ab1791, RRID:AB_302613 |
| Rabbit anti-RbAp48 Antibody | Abcam | Cat# ab79416, RRID:AB_1603754 |
| Rabbit anti-MYC Antibody | Abcam | Cat# ab32072, RRID:AB_731658 |
| Mouse anti-MYC Antibody | Santa Cruz | Cat# sc-40, RRID:AB_627268 |
| Rabbit anti-NCoR Antibody | Abcam | Cat# ab24552, RRID:AB_2149005 |
| Rabbit anti-MBD2 Antibody | Abcam | Cat#ab188474 |
| Rabbit anti-MBD3 Antibody | Abcam | Cat#ab157464 |
| Rabbit anti-CHD3 Antibody | Abcam | Cat# ab109195, RRID:AB_10862514 |
| Rabbit anti-p21 Antibody | Abcam | Cat# ab109199, RRID:AB_10861551 |
| Rabbit anti-GAPDH Antibody | Abcam | Cat# ab9485, RRID:AB_307275 |
| Rabbit anti-RBP1 Antibody | Abcam | Cat# ab154881, RRID:AB_2721835 |
| Mouse anti-H3K27cr Antibody | PTM Biolabs | Cat#PTM-Bio 526 |
| Mouse anti-GFP Antibody | CMCTAG | Cat#AT0028 |
| Rabbit anti-GFP Antibody | CMCTAG | Cat#AT0044 |
| Mouse anti-Flag Antibody | CMCTAG | Cat#AT0022 |
| Rabbit anti-Flag Antibody | CMCTAG | Cat#AT0502 |
| Rabbit anti-c-Myc tag Antibody | CMCTAG | Cat#AT0045 |
| Mouse anti-c-Myc tag Antibody | CMCTAG | Cat#AT0023 |
| Mouse anti 6xHis-tag Antibody | BBI Life Sciences | Cat#D191001 |
| Rabbit anti-SIN3A Antibody | Cell Signaling Technologies | Cat# 8056, RRID:AB_11217624 |
| Rabbit anti-H3K27me3 Antibody | Cell Signaling Technologies | Cat# 9733, RRID:AB_2616029 |
| Normal rabbit IgG | Cell Signaling Technologies | Cat# 2729, RRID:AB_1031062 |
| Normal mouse IgG | Santa Cruz | Cat# sc-2025, RRID:AB_737182 |
| Rabbitanti-HDAC1 Antibody | Active motif | Cat# 40967, RRID:AB_2614948 |
| Rabbit anti-HDAC2 Antibody | Proteintech | Cat# 16152-1-AP, RRID:AB_2118514 |
| Mouse anti-alpha-tubulin Antibody | Proteintech | Cat# 66031-1-Ig, RRID:AB_11042766 |
| Mouseanti-GAS41 Antibody | Santa Cruz | Cat# sc-393708, RRID:AB_2892567 |
| Rabbit anti-GAS41 Antibody | Genetex | Cat# GTX16452, RRID:AB_2887538 |
| Mouseanti-Actin Antibody | Beyotime | Cat# AA128, RRID:AB_2861213 |
| Goat anti-Mouse IgG H&L (HRP) 2ND Antibody | Abcam | Cat# ab97040, RRID:AB_10698223 |
| Goat anti-Rabbit IgG H&L (HRP) 2ND Antibody | Abcam | Cat# ab97080, RRID:AB_10679808 |
| Mouse anti-FlagAntibody | Sigma | Cat# F1804, RRID:AB_262044 |
| Rabbit anti H2A.Z Antibody | Cell Signaling Technologies | Cat# 50722, RRID:AB_2799379 |
| Rabbit anti TIP60 Antibody | Protein tech | Cat# 10827-1-AP, RRID:AB_2128431 |
| Bacterial and virus strains | ||
| E. coli BL21(DE3) | TransGen Biotech | Cat# CD601 |
| E. coli DH5alpha | TransGen Biotech | Cat# CD201-02 |
| E. coli RFzero | Mukai et al. (2011)60 | N/A |
| E. coli JM109 (DE3) | Promega | P980A |
| Biological Samples | ||
| Human colorectal cancer cDNA microarray | OutdoBiotech | Cat# cDNA-HColA060CS02 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Protease inhibitor Cocktail | Roche | Cat# 11873580001 |
| Phusion High-Fidelity DNA Polymerase | ThermoFisher Scientific | Cat# F530S |
| RNAiMAX Transfection Reagent | Invitrogen | Cat# 13778150 |
| TRIzol reagent | Invitrogen | Cat# 15596026 |
| Doxorubicin | Selleck | Cat# S1208 |
| Idarubicin | Selleck | Cat# S1228 |
| Nutlin-3a | Selleck | Cat# S8059 |
| Blasticidin S HCl | Selleck | Cat# S7419 |
| Bezonase Nuclease | Sigma | Cat# E1014-25KU |
| Crotonyl-CoA | Sigma | Cat. 28007, lot BCBZ4793 |
| Dynabeads® Protein G | Invitrogen | Cat# 10004D |
| Straptavidin Magnetic beads | Engibody | Cat# IF9042 |
| GFP-Trap | ChromoTek | Cat# GTM-20 |
| H-Lys(Ac)-OH | BACHEM | Cat# 4015284 |
| H-Lys(crotonyl)-OH | BLDpharm | Cat# BD328520 |
| Synthesized H3 K27cr (a.a. 21-33) peptide (ATKAAR-Kcr-SAPATG) | This paper | Mimotopies |
| Synthesized H3 K27ac (a.a. 21-33) peptide (ATKAAR-Kac-SAPATG) | This paper | Mimotopies |
| Synthesized H3 K27 (a.a. 21-33) peptide (ATKAARKSAPATG) | This paper | Mimotopies |
| Synthesized H3 K9cr (a.a. 3-15) peptide (TKQTAR-Kcr-STGGKA) | This paper | Mimotopies |
| Synthesized H3 K9ac (a.a. 3-15) peptide (TKQTAR-Kac-STGGKA) | This paper | Mimotopies |
| Synthesized H3 K18cr (a.a. 12-24) peptide (GGKAPR-Kcr-QLATKA) | This paper | Mimotopies |
| Synthesized H3 K18ac (a.a. 12-24) peptide (GGKAPR-Kac-QLATKA) | This paper | Mimotopies |
| Synthesized H3 (a.a.1-25) peptide (ARTKQTARKSTGGKAPRKQLATKAA) | This paper | Mimotopies |
| Synthesized H3 K14cr (a.a.1-25) peptide (ARTKQTARKSTGG-Kcr-APRKQLATKAA) | This paper | Mimotopies |
| Synthesized H3 K14ac (a.a.1-25) peptide (ARTKQTARKSTGG-Kac-APRKQLATKAA) | This paper | Mimotopies |
| Recombinant protein YEATS4 (a.a. 11-150) | This paper | N/A |
| Recombinant protein YEATS4 (a.a. 11-165) | This paper | N/A |
| Recombinant protein YETAS4 (a.a. 11-150) Y74A/W93A | This paper | N/A |
| Recombinant protein c-Myc (a.a. 1-262) | This paper | N/A |
| Recombinant protein mGFP YEATS4 | This paper | N/A |
| Critical Commercial Assays | ||
| Re-ChIP-IT | Active Motif | Cat# 53016 |
| Streptavidin-coated 96-well plate | Thermo Scientific | Cat# 15501 |
| QuickChange Site-Directed Mutagenesis Kit | Agilent Technologies | Cat# 200518 |
| Power SYBR green PCR master mix | Applied Biosystems | Cat# A25778 |
| EasyScript One-Step gDNA Removal and cDNA Synthesis SuperMix | Transgen | Cat# AE311-03 |
| Plasmid Plus Midi Kit | Qiagen | Cat# 12943 |
| Deposited Data | ||
| GAS41 YEATS domain/H3K27cr structure | This paper | PDB:8I60 |
| RNA-seq and ChIP-seq data | This paper | GSE128590 |
| Western blot and cell fluorescence images | This paper | Mendeley Data: https://dol.org/10.17632/wn5dgnz64x.1 |
| Experimental Models: Cell Lines | ||
| HEK293T | COBIOER | Cat# CBP60439 |
| HCT116 | COBIOER | Cat# CBP60028 |
| HEK293T GAS41 KO | This paper | N/A |
| HEK293T MYC KO | This paper | N/A |
| HEK293T HDAC1 KO | This paper | N/A |
| HEK293T SIN3A KO | This paper | N/A |
| HCT116 GAS41 KO | This paper | N/A |
| HCT116 GAS41 KO stably expressing GFP-Flag-GAS41 | This paper | N/A |
| HCT116 GAS41 KO stably expressing GFP-Flag-GAS41 Y74A&W93A | This paper | N/A |
| HCT116 GAS41 KO stably expressing GFP-Flag vector | This paper | N/A |
| HCT116 GAS41 KO stably expressing 3xFlag-GAS41 | This paper | N/A |
| HCT116 GAS41 KO stably expressing 3xFlag-GAS41 H43S | This paper | N/A |
| HCT116 GAS41 KO stably expressing 3xFlag-vector | This paper | N/A |
| Experimental Models: Organisms/Strains | ||
| Mouse | Taconic, Germantown, NY | NOD-SCID, female, 6-8 weeks old. |
| Oligonulceotides | ||
| Oligonucleotides used in this study | This paper | Table S4 |
| Recombinant DNA | ||
| Modified pET32a_GAS41 (a.a. 11-150) | This paper | N/A |
| Modified pET32a_GAS41 (a.a. 11-150) W93A | This paper | N/A |
| Modified pET32a_GAS41 (a.a. 11-150) Y74A/W93A | This paper | N/A |
| Modified pET32a_GAS41 (a.a. 11-150) Y139A | This paper | N/A |
| Modified pET32a_GAS41 (a.a. 11-150) del 123-128 | This paper | N/A |
| Modified pET32a_GAS41 (a.a. 11-165) | This paper | N/A |
| Modified_pET32a_MYC (a.a. 1-262) | This paper | N/A |
| pET21a_GAS41 (a.a. 11-150) | This paper | N/A |
| Modified pET15b_hH2A type 1-B/E | This paper | N/A |
| Modified pET15b_hH2B type 1-J | This paper | N/A |
| Modified pET15b_hH3.1 | This paper | N/A |
| pCR2.1 TOPO_hH3.1_K27amber | This paper | N/A |
| Modified pET15b_hH4 | This paper | N/A |
| Modified pET24_KacRS_6mt | Wakamori et al. (2015)61 | N/A |
| Modified pET15b_Kcr-RS | This paper | N/A |
| Modified pUC_one-half of a 147-bp palindromic human α-satellite DNA | Davey et al. (2002)62 | N/A |
| pRK793: TEV protease | Kapust et al. (2001)63 | Addgene Plasmid #8827 |
| CbF_Flag-GAS41 | Robert G. Roeder | N/A |
| CbF-vector | This paper | N/A |
| CbF_Flag-GAS41 N (a.a.1-165) | This paper | N/A |
| CbF_Flag-GAS41 C (a.a.148-227) | This paper | N/A |
| CbF_Flag-GAS41 del 71-75aa | This paper | N/A |
| CbF_Flag-GAS41 Y74A | This paper | N/A |
| CbF_Flag-GAS41 W93A | This paper | N/A |
| CbF_Flag-GAS41 Y74A&W93A | This paper | N/A |
| CbF_Flag-GAS41 H43A | This paper | N/A |
| CbF_Flag-GAS41 H43S | This paper | N/A |
| CbF_Flag-GAS41 Y139A | This paper | N/A |
| CbF-myc-SIN3A | This paper | N/A |
| pEGFP-C1 | Clonetech | Cat#6084-1 |
| pEGFP-MYC | This paper | N/A |
| pEGFP-MYC MBI | This paper | N/A |
| pEGFP-MYC MBII | This paper | N/A |
| pEGFP-MYC MBIII | This paper | N/A |
| pEGFP-MYC MBII+III | This paper | N/A |
| pEGFP-MYC MBIV+BZ | This paper | N/A |
| pEGFP-GAS41 | This paper | N/A |
| pCAG-GFP-Flag vector | This paper | N/A |
| pCAG-GFP-Flag-GAS41 | This paper | N/A |
| pCAG-GFP-Flag-GAS41 Y74A&W93A | This paper | N/A |
| pCAG-GFP-HDAC1 | This paper | N/A |
| pCDNA3.1(−) | ThermoFisher Scientific | Cat#V79520 |
| pcDNA3.1-myc-MYC | This paper | N/A |
| pcDNA3.1-Flag-MYC | This paper | N/A |
| pcDNA3.1-Flag-HDAC1 | This paper | N/A |
| pCAG-3xFlag vector | This paper | N/A |
| pCAG-3xFlag-GAS41 | This paper | N/A |
| pCAG-3xFlag-GAS41 H43S | This paper | N/A |
| pCAG-3xFlag-GAS41 Y139A | This paper | N/A |
| pSpCas9(BB)-2A-GFP | Addgene: PX458 | Addgene |
| Software and Algorithms | ||
| FlowJo | FLOWJO, LLC | https://www.flowjo.com |
| Image J | NIH | https://imagej.nih.gov/ij/ |
| ModFit LT | ModFit LT | https://modfit-lt.software.informer.com/ |
| Prism 8 | GraphPad | https://www.graphpad.com |
| Origin 7.0 | OriginLab | https://www.originlab.com |
| Igor Pro 8 | WaveMetrics | https://www.wavemetrics.com/ |
| Modeller (v. 9.16) | Webb & Sali (2016)64 | https://salilab.org/ |
| iMOSFLM | Battye et al. (2011)65 | https://www.mrc-lmb.cam.ac.uk/mosflm/imosflm/ver740/introduction.html |
| BALBES | Long et al. (2008)66 | https://www2.mrc-lmb.cam.ac.uk/groups/murshudov/content/balbes/balbes_layout.html |
| Phenix.refinement | Afonine et al. (2012)67 | https://phenix-online.org/ |
| COOT | Emsley et al. (2010)68 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| PyMOL | Schrödinger, LLC | https://pymol.org/2/ |
| NMRPipe | Delaglio et al. (1995)69 | https://www.ibbr.umd.edu/nmrpipe/install.html |
| NMRViewJ | Johnson & Blevins (1994)70 | https://nmrfx.org/nmrfx/nmrviewj |
| HDExaminer | Sierra Analytics | https://www.leaptec.com/ |
| MassLynx 4.2 | Waters | https://www.waters.com/nextgen/us/en.html |
| Trimmomatic v0.36 | Bolger et al. (2014)71 | http://www.usadellab.org/cms/?page=trimmomatic |
| Bowtie v1.0.1 | Langmead et al. (2009)72 | http://bowtie-bio.sourceforge.net/index.shtml |
| IGV tools v2.3.32 | Robinson et al. (2011)73 | http://www.broadinstitute.org/igv/ |
| MACS v2.1.1 | Zhang et al. (2008)74 | https://macs3-project.github.io/MACS/ |
| Bedtools v2.21.0 | Quinlan et al. (2010)75 | https://bedtools.readthedocs.io/en/latest/index.html |
| DESeq2 v1.16.1 | Love et al. (2014)76 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| TopHat v2.1.0 | Kim et al. (2013)77 | https://anaconda.org/bioconda/tophat |
| HT-seq v0.6.0 | Anders et al. (2014)78 | https://htseq.readthedocs.io/en/master/index.html |
| Other | ||
| HisTrap™ FF column, 5ml | GE Healthcare | Cat#17-0921-02 |
| HisTrap™ HP column, 5ml | GE Healthcare | Cat#17-5247-01 |
| Superdex 75 26/60 | GE Healthcare | Cat#28-9893-34 |
| Superdex 200 16/60 | GE Healthcare | Cat#28-9893-35 |
| Superdex 200 Increase 10/300L GL | GE Healthcare | Cat# 28-9909-44 |