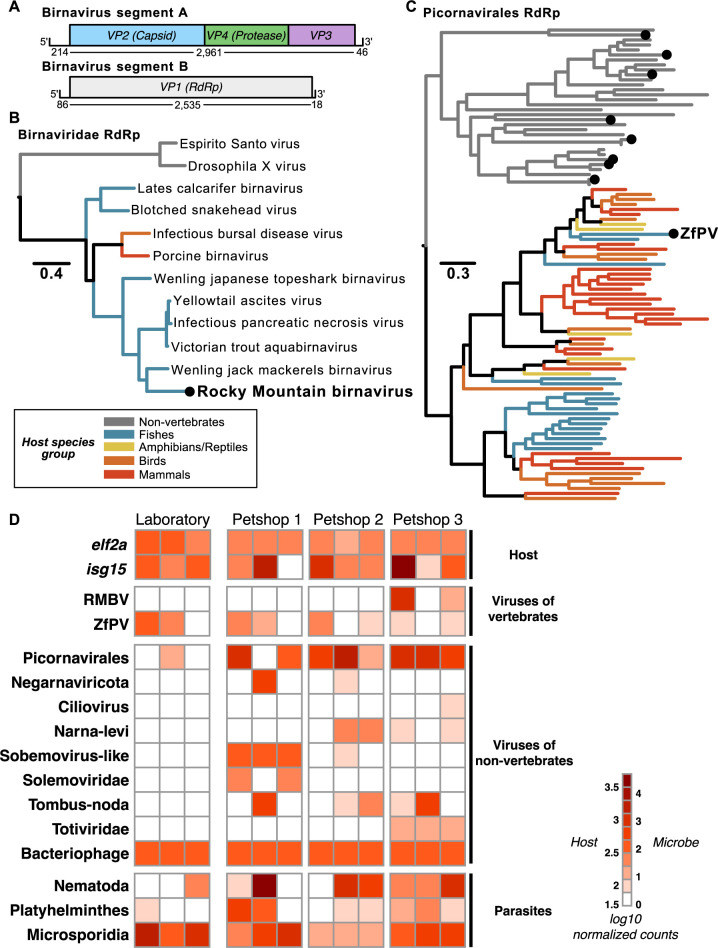
Fig 1. Metatranscriptomic survey of microbes associated with zebrafish from laboratory and pet trade sources.
. (A) Schematic representation of a newly discovered birnavirus associated with zebrafish in this study. Both segments of the linear dsRNA genome are depicted with boxes denoting relative positions of ORFs based on protein domain conservation. Numbers beneath segments indicate nucleotide lengths of each prospective 5′ UTR, coding sequence, and 3′ UTR. (B) Maximum likelihood phylogeny of viruses in the family Birnaviridae. The black dot highlights the novel zebrafish-associated virus schematized in (A), which we name RMBV. (C) Maximum likelihood phylogeny of RdRp protein sequences from viruses in the order Picornavirales. Black dots highlight the zebrafish-associated viruses that were detected (ZfPV) or newly discovered in this study. Trees in (A and C) are midpoint rooted. Scales are in amino acid substitutions per site. Branches are colored based on the known or inferred host group. (D) Taxonomic classification and quantification of host and microbe transcripts identified in intestinal tissues of zebrafish from laboratory and pet trade sources. Columns show estimated abundances in tissue samples from independent individuals grouped by source. RdRp, RNA-dependent RNA polymerase; RMBV, Rocky Mountain birnavirus; ZfPV, zebrafish picornavirus.