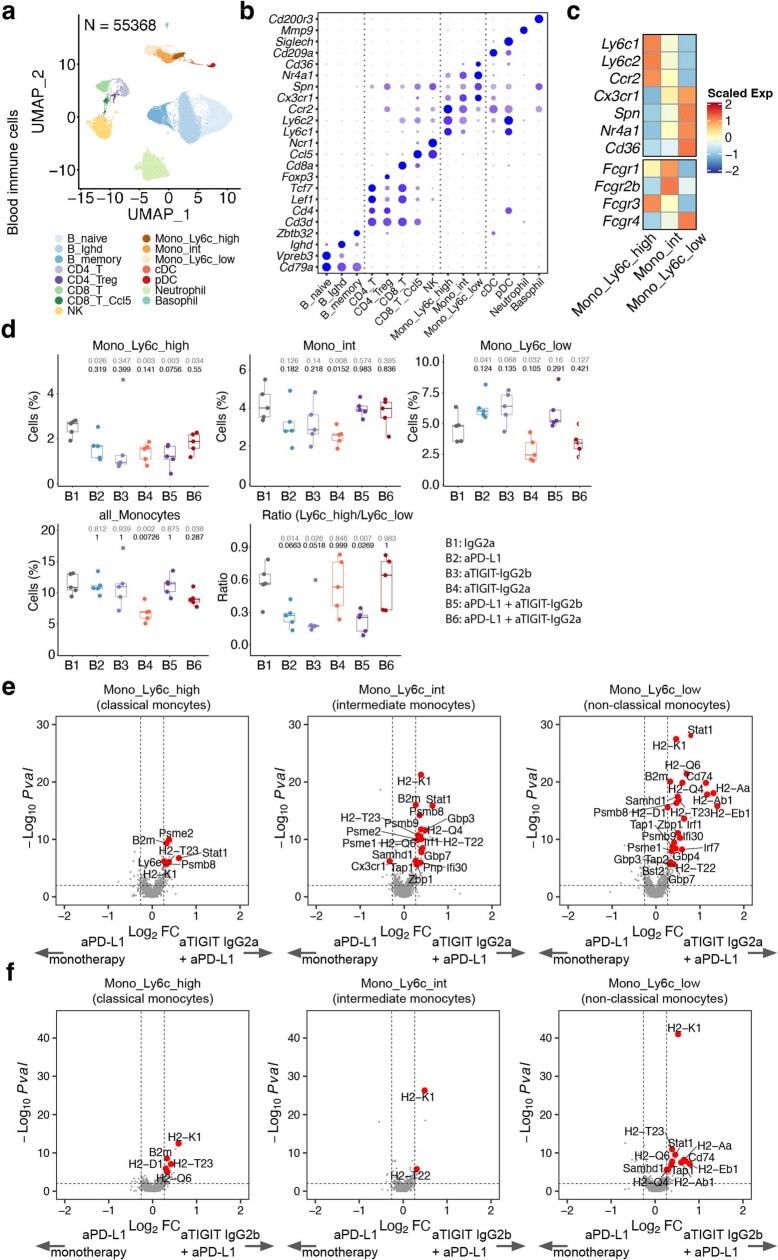
Extended Data Fig. 7. The modulation effects of anti-PD-L1 + anti-TIGIT on peripheral blood monocytes depends on the anti-TIGIT mAb Fc region.
a, UMAP of single cells from the peripheral blood cells (n = 55,368) coloured by cell types. b, Bubble plot showing the marker gene expression of cell types as in (a). c, Heatmap displaying the scaled gene expression of marker genes distinguishing classical, non-classical, and intermediate monocytes, and the expression patterns of FcɣR. d, Box plots comparing cell proportions of different treatments versus IgG2a isotype control (B1). B2, aPD-L1; B3, aTIGIT-IgG2b; B4, aTIGIT-IgG2a; B5, aPD-L1 + aTIGIT-IgG2b; B6, aPD-L1 + aTIGIT-IgG2a. Boxplot center line, median; box, interquartile range (IQR; the range between the 25th and 75th percentile); whiskers, 1.58 × IQR. Normal P values by two-tailed unpaired Student’s t-test are shown in grey colour; adjusted P values by Dunnett’s multiple comparison were shown in black colour. e, f, Volcano plots showing the gene expression of anti-PD-L1 + anti-TIGIT IgG2a versus anti-PD-L1 (e), and anti-PD-L1 + anti-TIGIT-IgG2b versus anti-PD-L1 (f) in peripheral blood classical (left), intermediate (middle), and non-classical (right) monocytes. P values were calculated by two-tailed Wilcoxon rank-sum test. a-f, Single cell RNA-seq was performed on peripheral CD45+ cells isolated at day 3 after treatment, and data are from one independent experiment with n = 5 mice in each group.