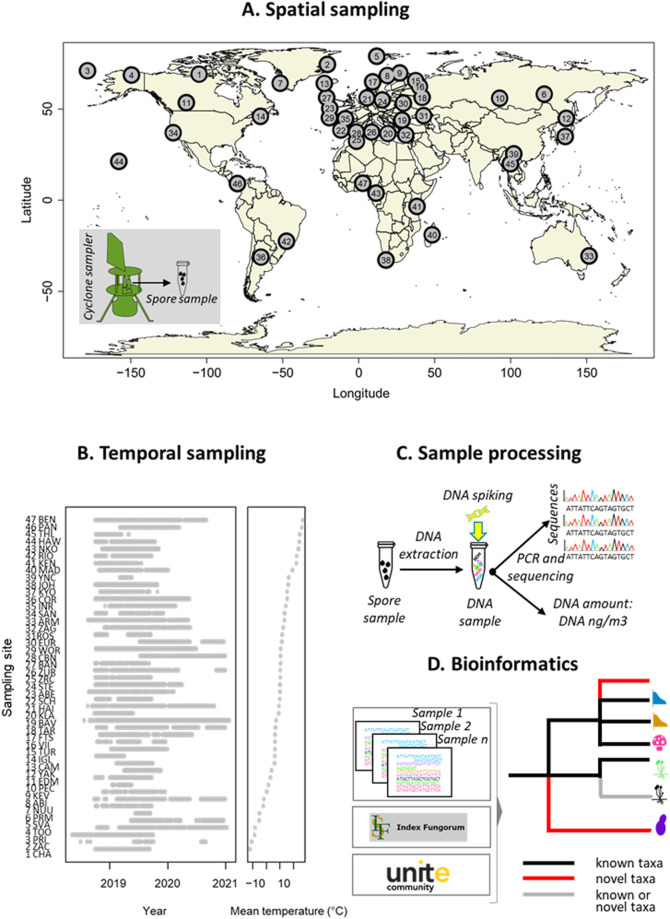
Fig. 1.
Study design and data generation pipeline of the Global Spore Sampling Project (GSSP). (A) The sampling design includes 47 sites with a global distribution, with the greatest coverage in Europe (22 sites) and the poorest coverage in the Southern hemisphere (6 sites). The airborne fungal samples were collected by a cyclone sampler, with each sample consisting of fungal spores filtered from 24 m3 of air during the 24-hr sampling period. (B) The study design included weekly samples for a sampling period over one to two years, with some variation among the sites caused mainly by logistical constraints. The sites are ordered according to their mean annual temperature. (C) We employed a metabarcoding approach to sequence the fungal ITS2 marker and quantified the amount of fungal DNA (in units of ng of DNA per m3 of air) using a spiking approach17. (D) We employed a bioinformatics pipeline that utilized denoising to obtain amplicon sequence variants (ASVs). We then combined probabilistic taxonomic placement with a constrained clustering approach to form species-level OTUs, and to place these OTUs in a taxonomic tree to the most resolved taxonomic level possible given the limitations of sequence reference databases. This tree consists of three types of branches: taxa that could be reliably assigned to previously known (black) and novel (red) taxa, and branches that may belong to either known or novel taxa (grey).